Fig. 1.
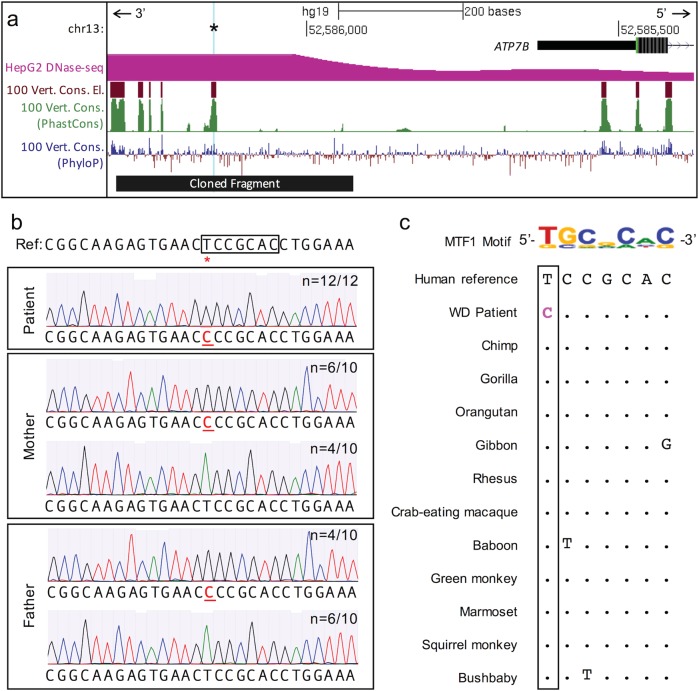
Genomic context of the candidate causative variant. a UCSC Genome Browser (hg19 minus strand) view of the region near the candidate causative variant positioned 676 bp upstream of the canonical ATP7B translation start site (chr13:g.52,586,149A>G, cyan highlight and asterisk). The variant lies in a 100-way vertebrate alignment conserved element and region of high cross-species sequence conservation as computed by PhastCons and PhyloP [30]. The chromatin surrounding chr13:g.52,586,149T>C is hypersensitive to DNaseI (and, therefore, open and accessible) in HepG2 cells. The sequence cloned into the luciferase reporter vector is indicated by the black bar. b Genotype verification for chr13:g.52,586,149T>C (red asterisk) by Sanger sequencing. Representative chromatograms show the indicated number (n=x/y) of amplification products containing the reference (T) or alternate (C) allele for each individual. Box indicates a predicted binding site for MTF1. c The candidate causative variant (bolded, pink) in the Wilson Disease (WD) patient disrupts a key base in the MTF1 position weight matrix. This exact base (boxed) is unaltered in all primates and in dozens of other mammals, even as distant as Tasmanian devil (Supplementary Fig. 1b) (color figure online).