Main conclusion
Our transient gene expression analyses in Arabidopsis protoplasts support the view that CK2αs and CK2βs positively and negatively modulate ABRE-dependent gene expression, respectively.
The phytohormone abscisic acid (ABA) regulates the expression of thousands of genes via ABA-responsive elements (ABREs), and has a crucial role in abiotic stress response. Casein kinase II (CK2), a conserved Ser/Thr protein kinase in eukaryotes, is essential for plant viability. Although the CK2 has been known as a tetrameric holoenzyme comprised of two catalytic α and two regulatory β subunits, each of the two types of subunits has been proposed to have independent functions. The Arabidopsis genome encodes four α subunits (CK2α1, CK2α2, CK2α3, CK2α4) and four β subunits (CK2β1, CK2β2, CK2β3, CK2β4). There is a growing body of evidence linking CK2 to ABA signaling and abiotic stress responses. However, the roles of each CK2 subunit in ABA signaling remain largely elusive. Using the transient expression system with the core ABA signaling components in Arabidopsis leaf mesophyll protoplasts, we show here that CK2α1 and CK2α2 (CK2α1/2) positively modulate ABRE-dependent gene expression as ABA signal output in ABA signaling, whereas all four CK2βs negatively modulate the ABRE-dependent gene expression mediated by subclass III SnRK2–AREB/ABF pathway and by CK2α1/2. These data indicate that CK2α1/2 and CK2βs positively and negatively modulate ABA signal output, respectively, suggesting that the quantitative balance of CK2 subunits determines the ABA signal output in plants. Given that CK2s act as pleiotropic enzymes involved in multiple developmental and stress–responsive processes, our findings suggest that CK2 subunits may be involved in integration and coordination of ABA-dependent and -independent signaling.
Electronic supplementary material
The online version of this article (10.1007/s00425-018-2919-5) contains supplementary material, which is available to authorized users.
Keywords: ABA-responsive-elements, Abscisic acid signaling, Arabidopsis thaliana, AREB/ABFs, Protoplast transient expression system, SnRK2
Introduction
The phytohormone abscisic acid (ABA) has crucial roles in a broad range of plant developmental processes and environmental stress responses (Cutler et al. 2010; Raghavendra et al. 2010; Miyakawa et al. 2013; Yoshida et al. 2015). Cellular dehydration during seed maturation and post-germination growth enhances endogenous ABA levels, which modulate the expression of many dehydration-responsive genes (Fujita et al. 2011). Approximately 10% of protein-coding genes in Arabidopsis thaliana plants are modulated by ABA, which is a much larger gene subset than that modulated by other phytohormones (Nemhauser et al. 2006). Many ABA-responsive genes carry conserved G-box-like cis-acting ABA-responsive elements (ABREs, PyACGTGG/TC) in their promoter regions (Mundy et al. 1990; Busk and Pages 1998; Hattori et al. 2002; Zhang et al. 2005; Gomez-Porras et al. 2007). ABA binds to the pyrabactin resistance1/PYR1-like/regulatory components of ABA receptor (PYR/PYL/RCAR) proteins to form ternary complexes with group-A protein phosphatase 2Cs (PP2Cs) (Ma et al. 2009; Miyazono et al. 2009; Nishimura et al. 2009; Park et al. 2009; Santiago et al. 2009). In the presence of ABA, the subclass III sucrose nonfermenting 1 related protein kinase 2 (SnRK2) is released from the PP2C-mediated negative regulation and phosphorylates basic leucine zipper (bZIP) transcription factors such as ABRE-binding protein/ABRE-binding factors (AREB/ABFs), which then activate the expression of ABA-responsive genes. Taken together with the recent results from a systemic study of a transcriptional network in response to ABA (Song et al. 2016), these findings indicate that ABRE-dependent gene expression constitutes the major ABA-responsive gene expression as ABA signal output in response to dehydration stress.
Casein kinase II (CK2) is a Ser/Thr protein kinase that is evolutionarily conserved in all eukaryotes. CK2 is essential for cell proliferation and survival, and is emerging as a potential target for anticancer pharmaceuticals (Mulekar and Huq 2014). CK2 is known to exist as a tetrameric holoenzyme composed of two catalytic α and two regulatory β subunits (Litchfield. 2003). On the other hand, several lines of evidence suggest that each of two types of subunit can exist independently and have independent functions as a monomer (Filhol et al. 2004). The Arabidopsis genome contains four α subunits (CK2α1, CK2α2, CK2α3, CK2α4) and four β subunits (CK2β1, CK2β2, CK2β3, CK2β4) genes. Recent molecular genetics and biochemical analyses suggest that CK2 is involved in ABA signaling and abiotic stress responses (for review: Vilela et al. 2015b). So far, studies of CK2 knockout mutants and RNAi lines suggest that CK2αs and CK2β1 are positive regulators of ABA responses (Mulekar et al. 2012; Mulekar and Huq 2014; Wang et al. 2014; Yuan et al. 2017), whereas biochemical studies suggest that CK2 negatively regulates SnRK2-mediated ABA signaling (Vilela et al. 2015a). It is thus controversial whether casein kinase II subunits (CK2s) positively or negatively regulate ABA signaling in plants. This discrepancy may be due to a lack of appropriate multiple knockout mutants of CK2α and CK2β required to determine the precise roles in ABA signaling, though CK2s have redundant functions as pleiotropic regulators of cell cycle, light signaling, circadian rhythms, flowering time, and hormone responses (Mulekar and Huq 2014). In fact, there are the adjacent positions of CK2α3 (At2g23080) and CK2α4 (At2g23070) on the chromosome, and ck2β2 and ck2β3 T-DNA insertion lines are not available. It is, therefore, challenging to determine the functions and roles of CK2s only by a genetic approach. In this study, to help overcome this problem, we used Arabidopsis leaf mesophyll protoplasts as a transient expression system to examine whether CK2s affect ABRE-dependent gene expression as an ABA signal output in ABA signaling. Here, we report that CK2αs positively modulate ABRE-dependent gene expression dependently and independently of the core ABA signaling pathway in the presence and the absence of exogenous ABA, respectively, whereas CK2βs negatively modulate ABRE-dependent gene expression in an exogenous ABA-independent manner.
Materials and methods
Plant materials and growth conditions
For transient expression analyses, A. thaliana L. accession Columbia-0 (Col-0, CS60000) and two triple knockout mutant lines areb1/2abf3 (Yoshida et al. 2010) and srk2d/e/i (Fujita et al. 2009; Nakashima et al. 2009) were grown in soil in pots (9.5 cm diameter) in an environmentally controlled chamber (CF-405S, TOMY, Osaka, Japan) at 22 °C under a 12-h light/12-h dark cycle (70 ± 20 μmol photons m−2 s−1). For transient expression analysis (cf. Fig. 2c), CS60000 were grown on GM agar plates to maintain high humidity as described previously (Fujita et al. 2005) with a 16-h light/8-h dark cycle (40 ± 10 µmol photons m−2 s−1). Similar results were obtained with these two methods.
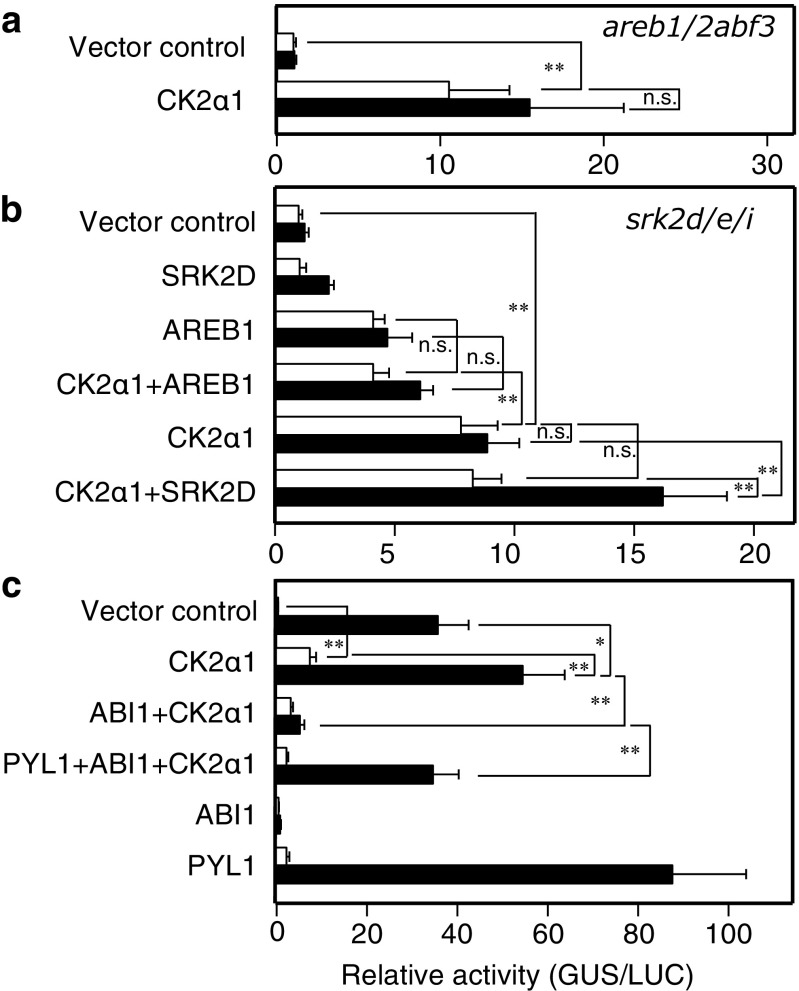
Fig. 2.
CK2α1 positively modulates ABRE-dependent gene expression independently and dependently of the core ABA signaling pathway in the absence and presence exogenous of ABA, respectively. a CK2α1 positively modulates ABRE-dependent gene expression in an AREB1/2ABF3-independent and -dependent manner in the absence and presence of exogenous ABA, respectively. Protoplasts were isolated from areb1/2abf3 triple mutant leaves. b CK2α1 positively modulates ABRE-dependent gene expression in a SnRK2D/E/I-independent and -dependent manner in the absence and presence of exogenous ABA, respectively. Protoplasts were isolated from srk2d/e/i triple mutant leaves. c PYL1 and ABI1 affect the stimulatory effect of CK2α1 on ABRE-dependent gene expression in the presence of exogenous ABA. Protoplasts were isolated from WT leaves. Open bars or filled bars indicate without ABA or with 2.0 μM ABA, respectively. Error bars indicate SD (n = 4). Experiments were performed at least three times, and a representative result is shown. *P < 0.05, **P < 0.01, n.s. no significant difference
Isolation of Arabidopsis leaf mesophyll protoplasts
Arabidopsis leaf mesophyll protoplasts were isolated as described previously (Yoo et al. 2007; Wu et al. 2009) with minor modifications. The leaves were collected from 2.5- to 4-week-old Arabidopsis plants. The upper epidermal leaf surfaces were affixed onto 19-mm width vinyl tape NO200-19-24 (Yamato, Tokyo, Japan). The basal epidermal leaf surfaces were affixed onto the same tape. The two strips of tape were then carefully torn away from each other to remove the lower epidermal cell layer. The peeled leaves attached to the tape were immersed in a 5 mL tube containing 4 mL of enzyme solution [20 mM Mes, pH 5.7, 1.5% (w/v) cellulase R10, 0.4% (w/v) macerozyme R10, 0.4 M mannitol and 20 mM KCl, and 10 mM CaCl2, 0.1% BSA]. Three tape strips (approximately 30 mm length per strip) were immersed in one tube. The tubes were gently shaken RT-50 (Taitec, Koshigaya, Japan) for 20–60 min. After the protoplasts were released into the solution, the tapes were removed from the solution and the protoplasts were collected by centrifuging at 100g for 2 min.
Transient expression analysis
Transient expression analysis was performed using protoplasts derived from Arabidopsis leaf mesophyll cells as described previously (Yoo et al. 2007) with minor modifications. Plasmid DNA was prepared using a plasmid DNA purification kit (Qiagen). The β-glucuronidase (GUS) reporter plasmid, RD29B-GUS (Uno et al. 2000), was co-transfected with effector plasmids and with the pBI35SΩ-ELUC (Mizoi et al. 2013) plasmid as an internal control to normalize protoplast transfection efficiency. The pBI35S-AREB1 plasmid (Fujita et al. 2005) was used as an effector plasmid. For analysis, RD29B-GUS (5.0 μg of plasmid per transfection) and pBI35SΩ-ELUC (1.0 μg per transfection) were used. Each transfection used 2.5 μg of effector plasmid, except for ABI1 (1.5 μg), AREB1 (1.0 μg) and CK2β (1.0 μg) in Fig. 3a, and SRK2D (0.625 μg) in Fig. 3b, per transfection. Total amounts of effector DNA per experiment, which include effector plasmids alone or combined with the vector control plasmid pSKX for transient expression analysis, were as follows: 2.5 μg (Fig. 1b), 5.0 μg (Figs. 2a, 3a, c), 6.5 μg (Fig. 2c), 7.5 μg (Fig. 2b), and 5.625 μg (Fig. 3b). ‘Relative activity’ indicates combined expression relative to the value obtained from the vector control. After transfection, protoplasts were incubated in 2 mL tubes at 22 °C for 14–18 h under dark conditions without ABA or with 2.0 μM ABA.
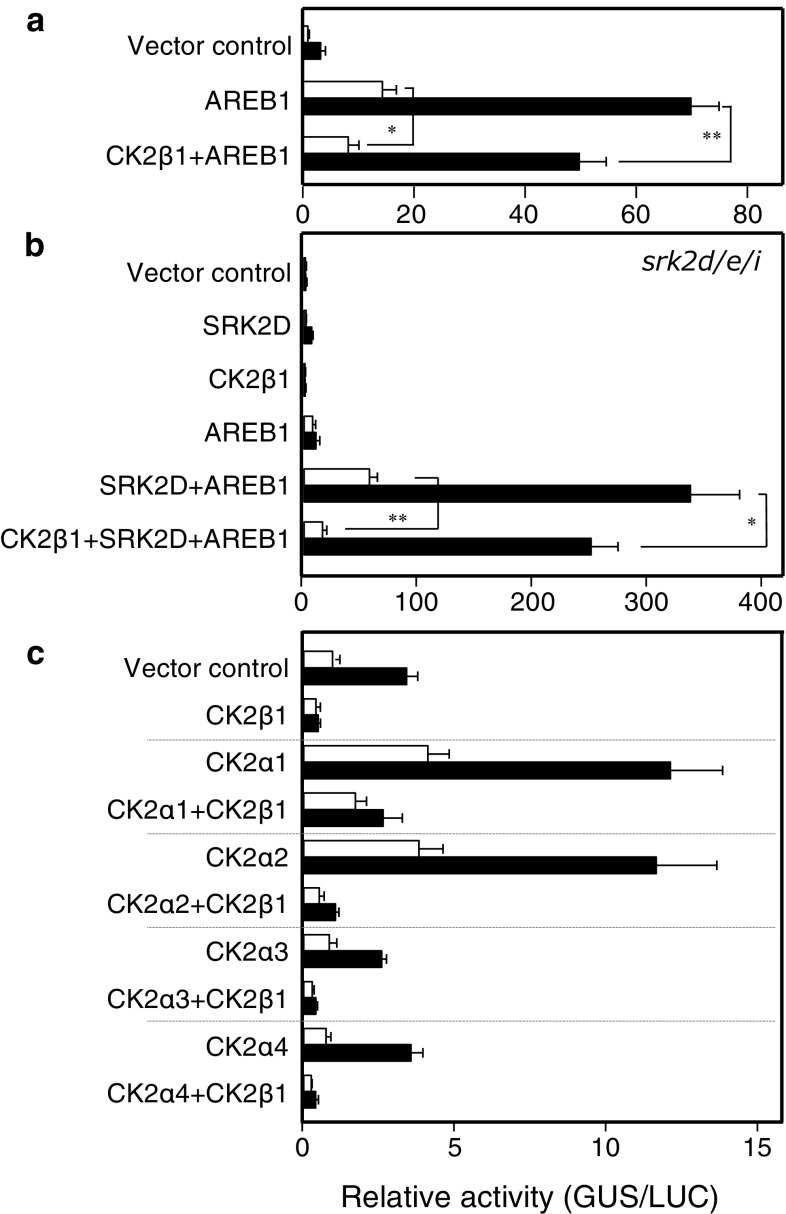
Fig. 3.
CK2β1 negatively modulates ABRE-dependent gene expression mediated by AREB–SnRK2 pathway and by CK2αs in an ABA-independent manner. a CK2β1 negatively modulates AREB1-mediated ABRE-dependent gene expression. Protoplasts were isolated from WT leaves. b CK2β1 negatively modulates AREB/SnRK2-mediated ABRE-dependent gene expression. Protoplasts were isolated from srk2d/e/i triple mutant leaves. c CK2β1 negatively modulates CK2α-mediated ABRE-dependent gene expression. Protoplasts were isolated from WT. Open bars or filled bars indicate without ABA or with 2.0 μM ABA, respectively. Error bars indicate SD (n = 4). Experiments were performed at least three times, and a representative result is shown. *P < 0.05, **P < 0.01, n.s. no significant difference
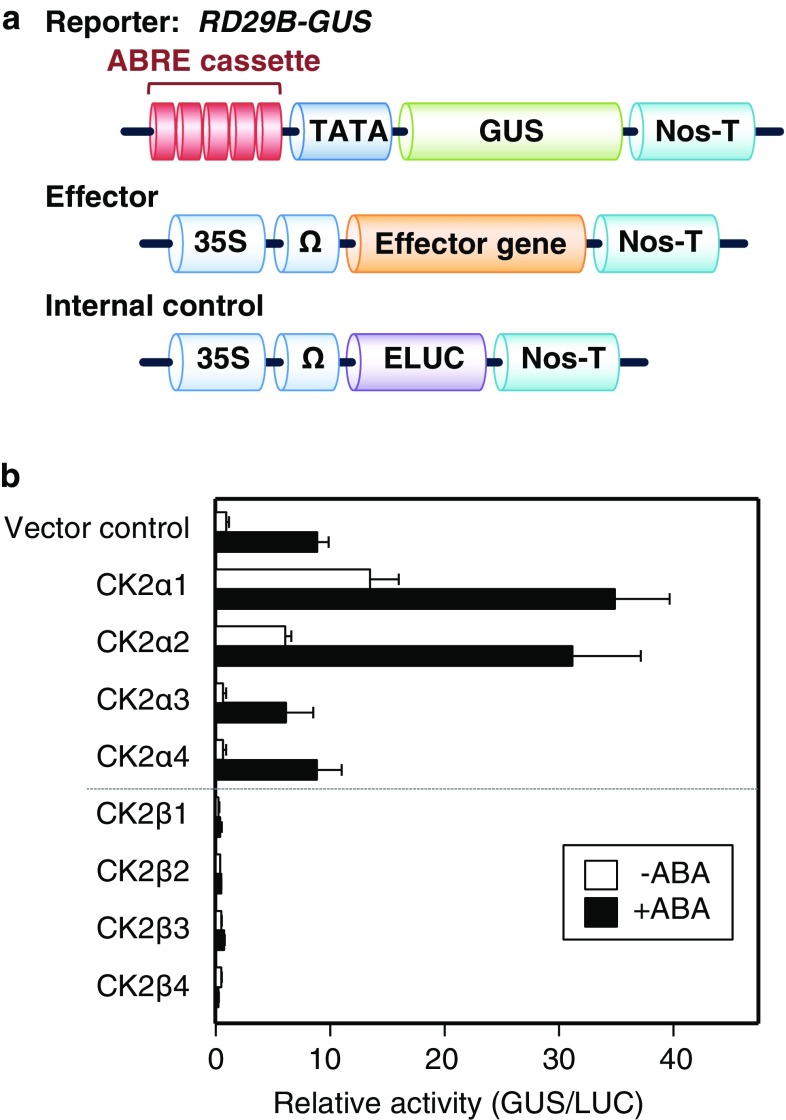
Fig. 1.
CK2α1/2 and CK2βs positively and negatively modulate ABRE-dependent gene expression in Arabidopsis leaf mesophyll protoplasts, respectively. a Scheme of reporter, effector, and internal control constructs used for transient expression analysis. The RD29B-GUS reporter construct carries five tandem copies of a 77-bp Arabidopsis RD29B promoter fragment containing two ABRE motifs fused to the GUS gene. The effector constructs contain the Cauliflower mosaic virus (CaMV) 35S promoter and tobacco mosaic virus Ω sequence fused to each cDNA fragment of interest. The pBI35SΩ-ELUC reporter construct was co-transfected as an internal control for transfection efficiency. b CK2α1/2 and CK2βs have inverse roles in ABRE-dependent gene expression in Arabidopsis leaf mesophyll protoplasts. Protoplasts were isolated from WT leaves. ‘Relative activity’ indicates combined expression relative to the value obtained from the vector control. Open bars or filled bars indicate without ABA or with 2.0 μM ABA, respectively. Error bars indicate SD (n = 4). Experiments were performed at least three times, and a representative result is shown
Plasmid construction for transient expression analysis
The primers used for DNA fragment amplification are listed in Supplementary Table S1.
pSKX
pSKX-based vectors were used for transient expression analysis of selected effectors, except for pBI35S-AREB1. A DNA fragment including El2-35SΩ-NosT was amplified by PCR from pGKX (Qin et al. 2008) using the forward and reverse PstI–HindIII linker primer pair (pGKXEl2-5′PstHind and pGrMCS-3′). The resulting PCR fragment was digested with PstI, subjected to end blunting and digested with KpnI, and inserted into the end-blunted SacI and KpnI site of pBluescriptIISK(−) to produce pSK-EI2-35SΩ (named pSKX).
pSKX-CK2αs and pSKX-CK2βs
For pSKX-CK2α1, pSKX-CK2α2, pSKX-CK2α3, pSKX-CK2α4, pSKX-CK2β1, pSKX-CK2β2, pSKX-CK2β3, and pSKX-CK2β4, DNA fragments of full-length open reading frames (FL ORFs) of CK2αs and CK2βs were PCR-amplified from their cDNA with BamHI–SacI linker primers. The resulting PCR products were digested with BamHI and SacI, and cloned into the BamHI and SacI sites of the plasmid vector pSKX.
pSKX-ABI1dMyc
The ABI1 FL ORF was PCR-amplified from its cDNA using ABI1F-NotI and ABI1R1-StuI primers, and the fragment was purified by ethanol precipitation. The resulting fragment was PCR-amplified using ABI1F-NotI and ABI1R4-dMyc-SmaI primers, purified, and further amplified using ABI1F-NotI and ABI1R5-dMyc-SmaI primers. The resulting PCR product was digested with NotI and SmaI, and cloned into the NotI and EcoRV sites of pSKX to produce pSKX-ABI1dMyc.
pSKX-SRK2DFlag
The FL ORF DNA of SRK2D was PCR-amplified from its cDNA using SRK2DF-NotIa and SRK2DR1-StuIa primers. The resulting fragment was purified by ethanol precipitation, and then subjected to further PCR amplification using SRK2DF-NotIa and SRK2DR2-Flag-EcoRV primers. The resulting PCR product was then digested with NotI and EcoRV, and cloned into the NotI and EcoRV sites of pSKX to produce pSKX-SRK2DFlag.
pSKX-HisPYL1
The FL ORF DNA of PYL1 was PCR-amplified from its cDNA using His-NcoI-PYL1F1a and PYL1R1-EcoRVa primers. The resulting fragment was purified by ethanol precipitation, and then subjected to further PCR amplification using NotI-His-NcoI-PYL1F2 and PYL1R1-EcoRVa primers. The resulting PCR product was digested with NotI and EcoRV, and cloned into the NotI and EcoRV sites of pSKX to produce pSKX-HisPYL1.
Phylogenetic analysis
Amino acid sequences were aligned and clustered using Clustal X2.1 (http://www.genome.jp/tools-bin/clustalw) (Larkin et al. 2007). The phylogenetic tree was constructed using MEGA7 (Kumar et al. 2016).
Results
CK2αs and CK2βs inversely modulate ABRE-dependent gene expression
ABRE-dependent gene expression plays a pivotal role in ABA-responsive gene expression as ABA signal output in response to dehydration stress (Fujita et al. 2011). Although there is a growing body of evidence linking CK2 to ABA signaling and abiotic stress responses (Vilela et al. 2015b), it remains unclear how CK2αs and CK2βs are involved in ABRE-dependent gene expression in ABA signaling. To examine the roles of all eight CK2s in ABA-responsive ABRE-dependent gene expression, we performed transient expression assays in Arabidopsis leaf mesophyll protoplasts using a β-glucuronidase (GUS) reporter gene, RD29B-GUS, driven by ABRE cis-elements derived from the ABA-responsive RD29B promoter (Uno et al. 2000) (Fig. 1a). Transfection of CK2α1 or CK2α2 induced RD29B-GUS expression in leaf mesophyll protoplasts in the presence and the absence of exogenous ABA compared with the vector control, whereas transfection of CK2α3 or CK2α4 did not (Fig. 1b). The findings are consistent with the observation that CK2α1 and CK2α2 are the closest homologs in the phylogenetic tree (Fig. S1), and only they have putative cleavage sites in the N-terminal region (Fig. S2). These collective data support the view that CK2α1 and CK2α2 (CK2α1/2) positively modulates ABRE-dependent gene expression (Fig. 1b). By contrast, CK2β transfection suppressed RD29B-GUS expression in leaf mesophyll protoplasts compared with the vector control in the presence and the absence of exogenous ABA, indicating that all four CK2βs negatively modulates ABRE-dependent gene expression in an ABA-independent manner (Fig. 1b). Collectively, these data indicate that CK2α1/2 and CK2βs positively and negatively modulate ABRE-dependent gene expression, respectively.
CK2α1 positively modulates ABRE-dependent gene expression
The core ABA signaling components (PYR/PYL/RCARs, group-A PP2Cs, subclass III SnRK2s, and AREB/ABF transcription factors) are necessary and sufficient for ABA perception, signaling, and ABA-responsive ABRE-dependent gene expression in a transient expression system based on Arabidopsis leaf mesophyll protoplasts (Fujii et al. 2009). To investigate how the CK2α1/2 positively modulates ABRE-dependent gene expression, we evaluated the effects of CK2α1 on RD29B-GUS expression in protoplasts derived from three different genotypes: an AREB/ABF subfamily triple mutant areb1/2abf3 (Yoshida et al. 2010), a subclass III SnRK2 subfamily triple mutant srk2d/e/i (Fujita et al. 2009; Nakashima et al. 2009), or wild-type (WT) Arabidopsis. Significant ABA-dependent induction of RD29B-GUS expression caused by transfection of CK2α1 could not be observed in areb1/2abf3 or srk2d/e/i protoplasts (Fig. 2a, b) unlike the transfection of CK2α1 in WT protoplasts (Figs. 1b, 2c), whereas co-transfection of SRK2D/SnRK2.2 (SRK2D) with CK2α1 recovered the ABA-dependent induction of RD29B-GUS expression in srk2d/e/i protoplasts (Fig. 2b). These data suggest that CK2α1 positively modulates ABRE-dependent gene expression in an AREB/SnRK2-dependent manner in the presence of ABA.
By contrast, even in the absence of exogenous ABA, transfection of CK2α1 alone induced significant level of RD29B-GUS expression in either areb1/2abf3, srk2d/e/i, or WT protoplasts compared with the vector control in each experiment (Figs. 1b, 2) and the co-transfection of CK2α1 with SRK2D did not affect RD29B-GUS expression in srk2d/e/i protoplasts compared with the transfection of CK2α1 alone in the srk2d/e/i protoplasts (Fig. 2b). These data support the notion that CK2α1 positively modulates ABRE-dependent gene expression in an AREB/SnRK2-independent manner in the absence of exogenous ABA. Indeed, in the absence of exogenous ABA in srk2d/e/i protoplasts, co-transfection of AREB1 with CK2α1 did not affect RD29B-GUS expression compared with the transfection of AREB1 alone (Fig. 2b), indicating that CK2α1 instead of SRK2D does not directly activate ABRE-dependent gene expression via AREB-mediated pathway in the absence of exogenous ABA. Moreover, in the absence of exogenous ABA in srk2d/e/i protoplasts, co-transfection of CK2α1 with AREB1 down-regulated RD29B-GUS expression compared with the transfection of CK2α1 alone (Fig. 2b), showing that the overexpression of AREB1 counteracts ABRE-dependent gene expression induced by CK2α1 in the absence of exogenous ABA. These data suggest that ABRE-binding transcription factors other than AREB/ABFs used in this study, which can compete with ABRE-dependent gene expression by AREB1, may be involved in CK2α1-mediated ABRE-dependent gene expression in the absence of exogenous ABA.
Next, we investigated the effects of CK2α1 combined with group-A PP2C and the PYR/PYL/RCAR ABA receptor on the induction of ABA-dependent induction of RD29B-GUS expression in WT protoplasts. As reported previously (Fujii et al. 2009), co-transfection of AREB1 with ABI1 inhibited RD29B-GUS expression in the presence of ABA, whereas co-transfection of PYL1 together with AREB1 and ABI1 partially recovered ABA-dependent induction of RD29B-GUS expression (Fig. S3). Co-transfection of CK2α1 with ABI1 inhibited RD29B-GUS expression, while co-transfection of CK2α1 together with ABI1 and PYL1 partially recovered ABA-dependent induction of RD29B-GUS expression (Fig. 2c). These data indicated that PYL1 and ABI1 affect the stimulatory effect of CK2α1 on ABRE-dependent gene expression in the presence of exogenous ABA. Together, our data support the hypothesis that CK2α1 positively modulates ABRE-dependent gene expression dependently of the core ABA signaling pathway in the presence of ABA.
CK2β1 negatively modulates ABRE-dependent gene expression mediated by AREB–SnRK2 pathway and by CK2αs
To elucidate the mechanism of CK2β-mediated suppression of ABRE-dependent gene expression, we evaluated the effects of CK2β1 co-transfection with AREB1 and/or SRK2D on ABA-mediated RD29B-GUS expression in srk2d/e/i and WT protoplasts. Co-transfection of CK2β1 with AREB1 suppressed RD29B-GUS expression that was induced by transfection of AREB1 alone with or without ABA treatment (Fig. 3a), indicating that CK2β1 negatively modulates AREB1-mediated ABRE-dependent gene expression. Co-transfection of CK2β1 with SRK2D and AREB1 attenuated RD29B-GUS expression that was induced by co-transfection of SRK2D and AREB1 in srk2d/e/i protoplasts with or without ABA treatment, indicating that the CK2β1 negatively modulates AREB/SnRK2-mediated ABRE-dependent gene expression (Fig. 3b). These results were consistent with a previous report that CK2 is involved in negative regulation of SnRK2 through enhancing SnRK2 interaction with PP2C and degradation (Vilela et al. 2015a). Finally, to determine whether CK2α and CK2β coordinately affect ABRE-dependent gene expression, we analyzed the effects of CK2β1 co-transfection with CK2αs on RD29B-GUS expression. Co-transfection of CK2β1 with CK2α1, CK2α2, CK2α3, or CK2α4 strongly suppressed RD29B-GUS expression compared with the transfection of CK2α1-4 alone with or without ABA treatment (Fig. 3c). These data indicated that CK2β1 negatively modulates CK2α-mediated ABRE-dependent gene expression.
Discussion
Here, we show that CK2α1/2 positively modulates ABRE-dependent gene expression dependently and independently of the core ABA signaling pathway in the presence and absence of exogenous ABA in Arabidopsis protoplasts, respectively (Figs. 1, 2). By contrast, the CK2βs negatively modulate ABRE-dependent gene expression mediated by AREB–SnRK2 pathway and by CK2αs in an exogenous ABA-independent manner in Arabidopsis protoplasts (Figs. 1, 3). These findings indicate that the CK2 α and β subunits inversely modulate ABRE-dependent gene expression as ABA signal output in Arabidopsis protoplasts, suggesting that the quantitative balance of CK2 subunits determines the ABA signal output in plants. However, it remains unclear whether the observed effects result from CK2 monomer function independently of holoenzyme or the interference with CK2 tetramer assembly. Given that the model proposed by Vilela et al. (2015a) in which CK2 negatively regulates ABA signaling through promoting SnRK2 degradation and enhancing SnRK2 interaction with PP2C, our data suggest a stimulatory effect of the catalytic subunits CK2α1 and CK2α2 which may be antagonized by the regulatory subunit CK2β1. Further research is required to clarify the regulatory mechanism of CK2 subunits in ABA signaling.
Our results show that in both the presence and absence of exogenous ABA, CK2s function as modulators of ABRE-dependent gene expression in Arabidopsis protoplasts (Figs. 1, 2, 3). This is in accordance with the previous findings that CK2 functions as a housekeeping gene (Mulekar and Huq 2014; Vilela et al. 2015b). Although so far CK2 has been identified as a negative regulator of ABA-activated SnRK2s in the core ABA signaling pathway (Vilela et al. 2015a), the roles and functions of CK2 subunits in the absence of exogenous ABA were unclear. We show here that CK2 α and β subunits positively and negatively modulate ABRE-dependent gene expression in an AREB/SnRK2-independent manner in the absence of exogenous ABA, respectively (Figs. 1, 2, 3). Since in the absence of exogenous ABA, CK2α1 instead of SnRK2 does not directly activate ABRE-dependent gene expression through AREB-mediated pathway, and the overexpression of AREB1 counteracts ABRE-dependent gene expression induced by CK2α1 (Fig. 2b), our collective data support the hypothesis that ABRE-binding transcription factors other than AREB/ABFs used in this study may activate ABRE-dependent gene expression downstream of CK2α1 in the absence of exogenous ABA. In contrast, considering the negative regulation model of CK2 in ABA signaling (Vilela et al. 2015a, b), the results suggest that negative modulation of ABRE-dependent gene expression by CK2β may be involved in ABRE/SnRK2-mediated pathway through the negative regulation of SnRK2 in the absence of exogenous ABA.
Thus, our analyses also provide the evidence that the protoplast transient expression system based on the ABRE-dependent gene expression is a useful tool to help overcome the problem in the limited genetic tools. On the basis of our results, combination studies of limited CK2s mutants with techniques of RNA silencing or CRISPR/Cas9, and transient expression analyses using the other marker genes, would provide more supportive information. Considering that CK2 acts as a pleiotropic enzyme involved in multiple developmental and stress–responsive processes and also functions as a housekeeping kinase regulating protein turnover in ABA signaling, the data presented here support the view that CK2 is a key modulator of crosstalk between ABA signaling and the other signaling pathways implicated in several other processes such as cell cycle, light signaling, and circadian rhythms. Together, this study suggests that CK2 subunits are involved in synergistically coordinating ABA-dependent and -independent signaling to modulate ABA signal output. Further works on CK2 interactors are needed to map novel CK2-mediated signaling networks that fine-tune ABA signal output in plants.
Author contribution statement
YN, MF, and YF designed the experiments and constructed the plasmids. YN performed the experiments and analyzed the data. YN and YF wrote the paper. YF conceived the research. All authors reviewed the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Table S1 Oligonucleotide primers used in this study. Suppl. Fig. S1 Phylogenetic tree of CK2αs and CK2βs in Arabidopsis. The neighbor-joining phylogenetic tree (Saitou and Nei 1987) was created using MEGA7 (Kumar et al. 2016). The optimal tree with the sum of branch length = 1.25324778 is shown. Bootstrap values (1000 replicates) are shown next to the branches (Felsenstein 1985). Evolutionary distances were computed using the p-distance method (Nei and Kumar 2000); units represent the number of amino acid differences per site. The subcellular localization of each CK2 is based on previous reports (Salinas et al. 2006; Perales et al. 2006; Portoles and Mas 2010: Mulekar and Huq 2015). CK2αs and CK2βs are shaded in pink and in yellow, respectively. Suppl. Fig. S2 Alignment of CK2 amino acid sequences in Arabidopsis. Comparison of amino acid sequences of CK2αs (a) and CK2βs (b). Conservation ratio at each site is shown by shading (black, 100%; gray, 75%). Red bars mark reported characteristic domains. Red arrow indicates a signal peptide cleavage site detected by SignalP 4.1 Server (Petersen et al. 2011). Suppl. Fig. S3 Reconstitution of ABA signaling pathway by co-transfection of AREB1, ABI1 and PYL1. Protoplasts were isolated from WT leaves. RD29B-GUS (5.0 μg of plasmid per transformation) and pBI35SΩ-ELUC (1.0 μg per transfection) were used as the ABA-responsive reporter and internal control, respectively. Each transfection used 2.5 μg of effector plasmid, except for ABI1, which used 1.5 μg per transfection. Total amounts of effector DNA were 6.5 μg, which include effector plasmids alone or combined with the vector control plasmid pSKX for transient expression analysis. ‘Relative activity’ indicates combined expression relative to the value obtained from the vector control. After transfection, protoplasts were incubated for 14-18 h under dark conditions without ABA (open bars) or with 2.0 μM ABA (filled bars). Error bars indicate SD (n = 4). Experiments were performed at least three times, and a representative result is shown (PDF 555 kb)
Acknowledgements
We thank U. Mitsuyasu, T. Yoshida, E. Ohgawara, K. Mogami, M. Kishimoto, N. Takano, R. Motohashi, I. Saito, and M. Ikegami for excellent technical support, M. Toyoshima for skillful editorial assistance, and T. Ogata for critical reading of the manuscript.
Abbreviations
- ABF
ABRE-binding factor
- ABRE
ABA-responsive element
- AREB
ABRE-binding protein
- CK2
Casein kinase II
- CK2s
Casein kinase II subunits
- PP2C
Group-A protein phosphatase 2C
- PYR1/PYL/RCAR
Pyrabactin resistance1/PYR1-like/regulatory components of ABA receptor
- SnRK2
Subclass III sucrose nonfermenting 1 related protein kinase 2
Funding
This research was supported in part by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan Grants-in-Aid for Scientific Research (C) (nos. 24510312 and 16K07412 to YF) and the Ministry of Agriculture, Forestry and Fisheries (MAFF) of Japan (to YN, YF).
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
Footnotes
The original version of this article was revised due to a retrospective Open Access order.
Change history
11/17/2018
The article Casein kinase 2 α and β subunits inversely modulate ABA signal output in Arabidopsis protoplasts, written by Yukari Nagatoshi, Miki Fujita, and Yasunari Fujita, was originally published electronically on the publisher’s internet portal (currently SpringerLink) on 24 May 2018 without open access. With the author(s)’ decision to opt for Open Choice the copyright of the article changed on 19 November 2018 to © The Author(s) 2018 and the article is forthwith distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
References
- Busk PK, Pages M. Regulation of abscisic acid-induced transcription. Plant Mol Biol. 1998;37:425–435. doi: 10.1023/A:1006058700720. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenesis: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Filhol O, Martiel JL, Cochet C. Protein kinase CK2: a new view of an old molecular complex. EMBO Rep. 2004;5:351–355. doi: 10.1038/sj.embor.7400115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, Kobayashi M, Nakasone S, Yamada K, Ito T, Shinozaki K, Yamaguchi-Shinozaki K. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- Gomez-Porras JL, Riano-Pachon DM, Dreyer I, Mayer JE, Mueller-Roeber B. Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genom. 2007;8:260. doi: 10.1186/1471-2164-8-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A. Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol. 2002;43:136–140. doi: 10.1093/pcp/pcf014. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Litchfield DW. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J. 2003;369:1–15. doi: 10.1042/bj20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Miyakawa T, Fujita Y, Yamaguchi-Shinozaki K, Tanokura M. Structure and function of abscisic acid receptors. Trends Plant Sci. 2013;18:259–266. doi: 10.1016/j.tplants.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, Yoshida T, Kodaira KS, Yamaguchi-Shinozaki K, Tanokura M. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- Mizoi J, Ohori T, Moriwaki T, Kidokoro S, Todaka D, Maruyama K, Kusakabe K, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. GmDREB2A;2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 2013;161:346–361. doi: 10.1104/pp.112.204875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulekar JJ, Huq E. Expanding roles of protein kinase CK2 in regulating plant growth and development. J Exp Bot. 2014;65:2883–2893. doi: 10.1093/jxb/ert401. [DOI] [PubMed] [Google Scholar]
- Mulekar JJ, Huq E. Arabidopsis casein kinase 2 alpha4 subunit regulates various developmental pathways in a functionally overlapping manner. Plant Sci. 2015;236:295–303. doi: 10.1016/j.plantsci.2015.04.013. [DOI] [PubMed] [Google Scholar]
- Mulekar JJ, Bu Q, Chen F, Huq E. Casein kinase II alpha subunits affect multiple developmental and stress–responsive pathways in Arabidopsis. Plant J. 2012;69:343–354. doi: 10.1111/j.1365-313X.2011.04794.x. [DOI] [PubMed] [Google Scholar]
- Mundy J, Yamaguchi-Shinozaki K, Chua NH. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc Natl Acad Sci USA. 1990;87:1406–1410. doi: 10.1073/pnas.87.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, Shinozaki K, Yamaguchi-Shinozaki K. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Portoles S, Mas P. The proteasome-dependent degradation of CKB4 is regulated by the Arabidopsis biological clock. Plant J. 2006;46:849–860. doi: 10.1111/j.1365-313X.2006.02744.x. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Portoles S, Mas P. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 2010;6:e1001201. doi: 10.1371/journal.pgen.1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress–responsive gene expression. Plant Cell. 2008;20:1693–1707. doi: 10.1105/tpc.107.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salinas P, Fuentes D, Vidal E, Jordana X, Echeverria M, Holuigue L. An extensive survey of CK2 alpha and beta subunits in Arabidopsis: multiple isoforms exhibit differential subcellular localization. Plant Cell Physiol. 2006;47:1295–1308. doi: 10.1093/pcp/pcj100. [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR. A transcription factor hierarchy defines an environmental stress response network. Science. 2016 doi: 10.1126/science.aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela B, Najar E, Lumbreras V, Leung J, Pages M. Casein kinase 2 negatively regulates abscisic acid-activated SnRK2s in the core abscisic acid-signaling module. Mol Plant. 2015;8:709–721. doi: 10.1016/j.molp.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Vilela B, Pages M, Riera M. Emerging roles of protein kinase CK2 in abscisic acid signaling. Front Plant Sci. 2015;6:966. doi: 10.3389/fpls.2015.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chang H, Hu S, Lu X, Yuan C, Zhang C, Wang P, Xiao W, Xiao L, Xue GP, Guo X. Plastid casein kinase 2 knockout reduces abscisic acid (ABA) sensitivity, thermotolerance, and expression of ABA- and heat–stress–responsive nuclear genes. J Exp Bot. 2014;65:4159–4175. doi: 10.1093/jxb/eru190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. Tape-Arabidopsis sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods. 2009;5:16. doi: 10.1186/1746-4811-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Mogami J, Yamaguchi-Shinozaki K. Omics approaches toward defining the comprehensive abscisic acid signaling network in plants. Plant Cell Physiol. 2015;56:1043–1052. doi: 10.1093/pcp/pcv060. [DOI] [PubMed] [Google Scholar]
- Yuan C, Ai J, Chang H, Xiao W, Liu L, Zhang C, He Z, Huang J, Li J, Guo X. CKB1 is involved in abscisic acid and gibberellic acid signaling to regulate stress responses in Arabidopsis thaliana. J Plant Res. 2017;130:587–598. doi: 10.1007/s10265-017-0924-6. [DOI] [PubMed] [Google Scholar]
- Zhang W, Ruan J, Ho TH, You Y, Yu T, Quatrano RS. Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress–responsive genes in Arabidopsis thaliana. Bioinformatics. 2005;21:3074–3081. doi: 10.1093/bioinformatics/bti490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Table S1 Oligonucleotide primers used in this study. Suppl. Fig. S1 Phylogenetic tree of CK2αs and CK2βs in Arabidopsis. The neighbor-joining phylogenetic tree (Saitou and Nei 1987) was created using MEGA7 (Kumar et al. 2016). The optimal tree with the sum of branch length = 1.25324778 is shown. Bootstrap values (1000 replicates) are shown next to the branches (Felsenstein 1985). Evolutionary distances were computed using the p-distance method (Nei and Kumar 2000); units represent the number of amino acid differences per site. The subcellular localization of each CK2 is based on previous reports (Salinas et al. 2006; Perales et al. 2006; Portoles and Mas 2010: Mulekar and Huq 2015). CK2αs and CK2βs are shaded in pink and in yellow, respectively. Suppl. Fig. S2 Alignment of CK2 amino acid sequences in Arabidopsis. Comparison of amino acid sequences of CK2αs (a) and CK2βs (b). Conservation ratio at each site is shown by shading (black, 100%; gray, 75%). Red bars mark reported characteristic domains. Red arrow indicates a signal peptide cleavage site detected by SignalP 4.1 Server (Petersen et al. 2011). Suppl. Fig. S3 Reconstitution of ABA signaling pathway by co-transfection of AREB1, ABI1 and PYL1. Protoplasts were isolated from WT leaves. RD29B-GUS (5.0 μg of plasmid per transformation) and pBI35SΩ-ELUC (1.0 μg per transfection) were used as the ABA-responsive reporter and internal control, respectively. Each transfection used 2.5 μg of effector plasmid, except for ABI1, which used 1.5 μg per transfection. Total amounts of effector DNA were 6.5 μg, which include effector plasmids alone or combined with the vector control plasmid pSKX for transient expression analysis. ‘Relative activity’ indicates combined expression relative to the value obtained from the vector control. After transfection, protoplasts were incubated for 14-18 h under dark conditions without ABA (open bars) or with 2.0 μM ABA (filled bars). Error bars indicate SD (n = 4). Experiments were performed at least three times, and a representative result is shown (PDF 555 kb)