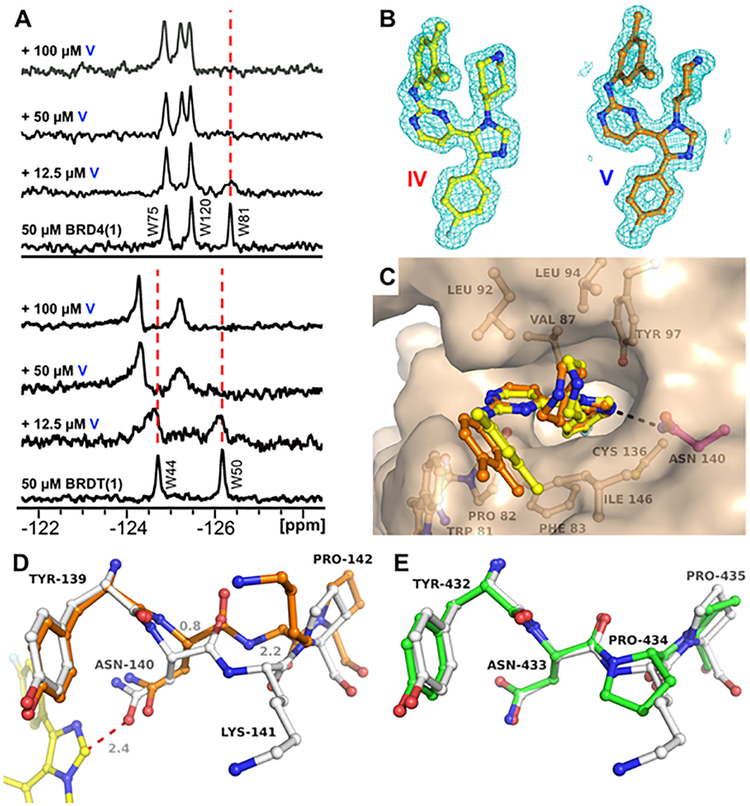
Figure 3.
Confirmation of compound V binding BRD4(1) and BRDT(1) and possible structural basis for the D1 over D2 selectivity of the inhibitors. (A) PrOF NMR titrations with bromodomains of BRD4 and BRDT. Samples contain 50 μM 19F-labeled BRD4(1) or BRDT(1) with increasing concentrations of ligand V. (B) Electron-density maps of ligands (2Fo − Fc, 1σ) from X-ray structures of BRD4(1) liganded with IV (yellow, 1.74 Å, PDB ID 6MH7) and V (orange, 1.60 Å, PDB ID 6MH1). (C) Superposition of compounds IV and V in the Kac site of BRD4(1) revealing subtle conformational changes of the dimethylphenyl moieties nested in the WPF shelf. (D) Unliganded state of BRD4(1) (gray, PDB ID 4IOR), with the Kac site around Asn140 in a relaxed state, and BRD4(1) bound to inhibitor V (yellow ligand and orange structure), with Asn140 giving way (Δ = 0.8 Å) as a result of steric hindrance with the imidazole moiety of the inhibitor (red dotted line) and positioning itself for optimal H-bonding interaction. As a result, the adjacent Lys141 undergoes a large conformational change (Δ = 2.2 Å). (E) Unliganded BRD4(2) (green, PDB ID 2OUO), with the region around Asn433 being highly similar to that of BRD4(1) (gray) except for the presence of Pro434 instead of lysine. The geometric constraints of the Asn−Pro−Pro sequence in BRD4(2) likely renders the peptide more rigid and less compatible with the inhibitor-induced conformational changes observed in BRD4(1).