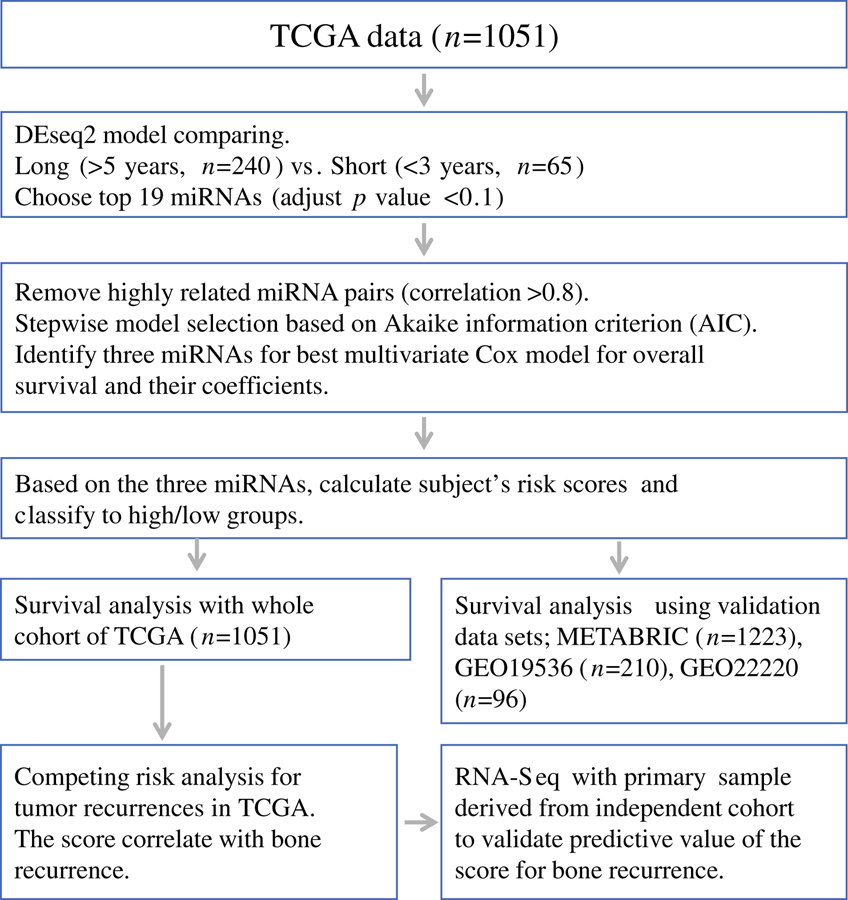
Figure 1.
Study strategy for selecting miRNA signature and generating risk scores to predict poor prognosis. Patients from two representative overall survival groups; “Long survival” (those survived greater than 5 years after diagnosis, n=240) vs. “Short survival” (deceased within 3 years of diagnosis, n=65); were used to identify the top miRNAs with differential expression using a model implemented in DEseq2 package based on the negative binomial distribution. First, we identified the top 19 miRNAs as our candidates, which showed most different expression levels in the two groups (adjust p-value <0.1). Next, highly related miRNA pairs (correlation>0.85) were excluded in order to reduce the multicollinearity and improve stability for further model selection. Finally, using stepwise model selection based on Akaike information criterion (AIC), we identified three miRNAs signature for best multivariate Cox proportional hazard model for overall survival and their coefficients (miR-19a, miR-93, and miR-106a). Calculated subject’s risk scores using three miRNAs signature and classify all patients of TCGA breast cancer into high score/low risk score groups. The same classification was made in the three independent validation datasets from METABRIC and GEO using these miRNAs.