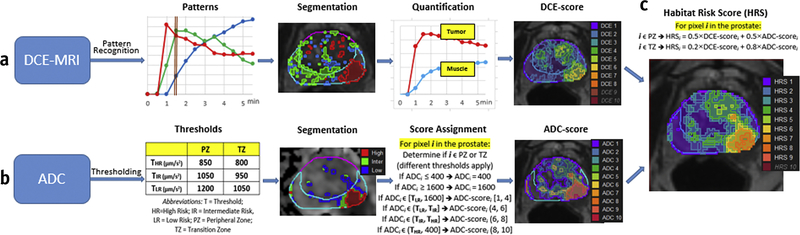
Figure 1. Flow-chart of Habitat Risk Score (HRS) construction.
(a) DCE-MRI analysis: </p>DCE-MRI data is baseline corrected by the average of δ = 2 pre-contrast points. Non Negative Matrix Factorization (NMF) is applied to the matrix D(X, t) with κ=3. D is represented as a product of κ basic temporal contrast signatures S(t) and their weights W(X), i.e. D ~W × S. The identified three basic temporal contrast signatures S(t) are shown in box, labelled Patterns; the well perfused pattern, Swp (depicted in red) is automatically selected as the pattern with maximum area under the curve (AUC) between 0 and 90 sec (brown bar). The segmented weights of the patterns W(t) are shown in corresponding colors in box, labelled Segmentation (α=60, β=10). The area in red depicts the region of interest (ROI) for the well-perfused pattern: ROIwp. Two curves are shown in box, labelled Quantification: the average DCE-curve from the ROIwp (Tumor) and Gluteus Maximus (Muscle). Semi-quantitative feature (Late AUC) is extracted from these two curves: Σi (Tumor) and Σim (Muscle). The ratio Σi/Σim is propagated for each pixel in the prostate, using Wwp. A value between 1 to 10 is assigned to each pixel of the prostate, using σ1=.5 and σ2=2.5 (the range is bound by the γth and (100-γ)th; γ=5). DCE-score is depicted as a heat map (Note that DCE9 and DCE10 are empty contours). (b) ADC analysis: Thresholds ΤHR, TIR and TLR for volumes at high risk, intermediate risk and low risk for cancer are identified separately for peripheral zone (PZ) and transition zone (TZ). The segmented volumes are shown in box, labelled Segmentation. The ADC is mapped on 10-point scale, using the rules in Score Assignment box and depicted as a heat map. (c) HRS is calculated for each pixel, using η1= 0.5 and η2= 0.5 in PZ and η1= 0.2 and η2 =0.8 in TZ. HRS is depicted as a heat map (Note that HRS10 is an empty contour).