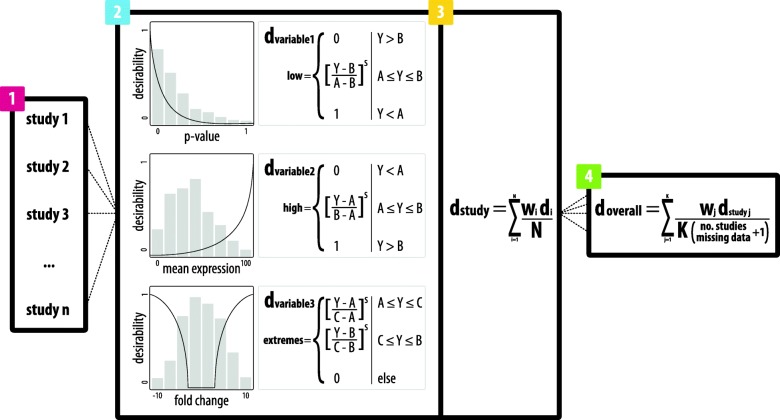
Fig. 2.
integRATE relies on three main steps to identify studies, integrate data, and rank candidate genes. (1) Relevant studies must first be identified for integration based on any number of features including, but not limited to: phenotype, experimental design, and data availability. (2) Data corresponding to all variables in each study are then transformed according to the appropriate desirability function. In this step, the user assigns a function based on whether low values are most desirable (dlow), high values are most desirable (dhigh), or extreme values are most desirable (dextreme) and can customize the shape of the function with other variables like cut points (A, B, C), scales (s), and weights (w) to better reflect the data distributions or to align with user opinion regarding data quality and relevance. (3) These variable-based scores are integrated (dstudy) with a straightforward arithmetic mean (where weights can also be applied) to produce a single desirability score for each gene in each study containing information from all variables simultaneously. (4) Finally, study-based desirability scores are integrated to produce a single desirability score for each gene (doverall) that includes information from all variables in all studies and reflects its cumulative weight of evidence from each data set identified in step 1. These scores are normalized by the number of studies containing data for each gene and can be used to rank and prioritize candidate genes for follow up computational and, most importantly, functional analyses