Abstract
Background
In the era of personalized cancer medicine, identifying techniques for effectively matching patients to efficacious treatments is a critical step in the treatment process. The advent of anti-cancer immunotherapies necessitates novel approaches to biomarker identification beyond traditional genomic profiling. One promising approach is incorporation of nomograms into treatment decisions. Nomograms are prediction tools, based on statistical modeling, designed to predict treatment outcomes. As a first step toward developing a nomogram, we conducted analyses to predict CD137 expression of natural killer cells after monoclonal antibody (mAb) treatment.
Patients and methods
Patient, tumor and immune characteristics were collected from 199 patients with breast cancer (N = 62), head/neck cancers (N = 46) and non-Hodgkin’s lymphoma (NHL) (N = 91), who were receiving mAb therapy as part of clinical trials. The difference in CD137 expression before and after mAb therapy was assessed by flow cytometry. To evaluate those who respond to mAb therapy via increased CD137 expression, we applied classification and regression trees (CART), multivariable lasso regression tools and Random Forest.
Results
The CD137 expression was significantly different for each cancer type [mean (SD): Breast: 6.6 (6.5); Head/Neck: 11.0 (7.0); NHL: 7.5 (7.1), P < 0.0001]. For breast cancer and NHL, FcR polymorphism and baseline CD137 expression were significant predictors of increased CD137 expression; for head/neck cancer, FcR polymorphism and age were significant predictors of increased expression.
Conclusions
Our preliminary results suggest that FcR polymorphism, pre-treatment CD137 expression and age are significant predictors of CD137 upregulation in patients. This study demonstrates that the development of a nomogram for therapy response is feasible. Further work validating our models in an independent cohort will provide the next steps in developing a nomogram that may be used to individualize this therapeutic approach for patients (NCT01114256).
Keywords: biomarker, CD137, natural killer cells, monoclonal antibody, cancer, regression tree
Key Messages
Proper patient stratification based on predictive biomarkers can improve outcome. We describe prognostic factors that predict CD137 expression on natural killer cells after treatment with monoclonal antibodies. Our efforts constitute the first step in developing a nomogram that predicts outcome.
Introduction
Recent clinical achievements of cancer immunotherapies have revolutionized cancer treatment, but with this renaissance in oncologic care come two potentially significant limitations: cost and toxicity. Rising health care costs and limited resources have raised concern over the affordability of novel immunotherapies. A prime example is the checkpoint inhibitor ipilimumab, with a cost of over $30 000 per administration in the USA [1, 2]. Concerns with toxicity have led to early termination of promising immunotherapies such as urelumab, an agonistic mAb that stimulates the tumor necrosis factor receptor superfamily member CD137, in 2009 [3, 4].
In 2012, anti-CD137 was re-launched following identification of therapeutic activity at lower doses without the limiting toxicity. Anti-CD137 is currently undergoing clinical evaluation in combination regimens with tumor-targeting mAbs in multiple types of cancer. Thus, the question we ask again is: how can we identify and select patients who may be predicted to derive significant benefit from anti-CD137, while not subjecting those predicted to obtain minimal to no benefit to the treatment, in order to reduce costs and toxicity and maximize efficacy?
Nomograms are graphical calculation instruments based on statistical predictive models that provide predictions on likely treatment outcomes [5–7]. Nomograms provide individualized risk estimates by incorporating and illustrating important prognostic factors, and are superior to other available decision aids in more accurately predicting outcomes in patients with cancer [8, 9].
We have identified CD137 as a marker of natural killer (NK) cell activation that is upregulated following ligation of the FcRγIII receptor (CD16) with the Fc portion of mAbs bound to the surface of tumor cells [10–12]. NK cells are important mediators of antibody dependent cell-mediated cytotoxicity (ADCC), one of the primary mechanisms of anti-tumor action of mAbs [13]. We have previously shown that agonistic anti-CD137 enhances mAb-mediated ADCC function and synergizes with anti-CD20, anti-HER2, and anti-EGFR in murine models of lymphoma, breast cancer, and colorectal cancer, respectively [10–12]. Our results provided a strong rationale for anti-CD137/mAb therapy combination regimens currently being evaluated in clinical trials.
As efficacy of anti-CD137 mAb is partially dependent upon expression of CD137 on NK cells, this has led to the speculation that CD137 could be used as a predictive biomarker of response to mAb therapy. In patients with non-Hodgkin’s lymphoma, breast cancer, and head and neck cancer, circulating NK cells upregulate CD137 expression within 24 h of receiving rituximab, trastuzumab, or cetuximab, respectively [10–12]. However, the observed increase in CD137 expression on NK cells is heterogeneous, with a subset of patients demonstrating minimal upregulation and others displaying a 4- to 5-fold increase.
Predicting CD137 upregulation based on patient clinicopathological data is the first step towards building a nomogram that utilizes CD137 as a biomarker of response to mAb therapies. If validated, it would provide clinicians with a powerful tool allowing selection and individualization of mAb therapies that optimizes patient responses while limiting toxicity and sparing resources. Understanding the magnitude of CD137 upregulation would also be helpful in selective screening of patients as candidates for clinical trials of anti-CD137 in combination with mAb therapies. We have taken the first step by characterizing the upregulation of CD137 on circulating NK cells in patients receiving mAb therapy in a prospective clinical trial. Our primary objective was to identify clinicopathological factors that can predict CD137 expression 24-h post-mAb therapies and to characterize the change in CD137 expression.
Methods
Study cohort and key data collected
We enrolled patients who met eligibility criteria at Stanford University Medical Center between 10 March 2010 and 1 January 2014. Eligibility criteria included the following: patients had histologically confirmed HER2+ breast adenocarcinoma (BC), squamous cell carcinoma of the head and neck (HNC), or CD20+ tumor (NHL); had not received any immunosuppressive or anti-cancer agent within 2 weeks prior to the first planned fine blood draw; and had a treatment plan that included a standard therapeutic mAb (trastuzumab, cetuximab or rituximab) administered on a schedule such that peripheral blood could be drawn immediately prior to and at 24 h after the first dose, with no cytotoxic chemotherapy in the interim.
All patients provided written informed consent in accordance with the Declaration of Helsinki after approval of the study by Stanford University’s Administrative Panels on Human Subjects (NCT01114256).
CD137 expression on circulating NK cells
Whole blood was collected in heparin tubes and processed within 24 h. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient separation using Ficoll-Paque PLUS (Amersham Biosciences) then stained for flow cytometric analysis using the following antibodies (BD Biosciences): CD3-PerCP, CD16-FITC, CD56-APC, and CD137-PE. Stained cells were collected on a FACSCalibur (BD Biosciences, San Jose, CA), and data were analyzed using Cytobank (http://www.cytobank.org).
Analysis of FcγRIIIa-158 polymorphisms
Genomic DNA was prepared from PBMCs using a DNA extraction kit (QIAGEN cat# 69504). Genotyping of FcγRIIIa-158 V/F polymorphism was performed by a polymerase chain reaction followed by allele-specific restriction enzyme digestion [14]. Genotyping was confirmed by direct sequencing of the region of interest.
Statistical analysis
Our primary outcome was post-mAb therapy CD137 expression. Our independent variables included all the demographic, pathologic, treatment and other cancer-specific characteristics as well as pre-mAb CD137 expression. Since, cancer type-specific variables were collected, each cancer type was evaluated separately. Our primary analysis was conducted using Classification and Regression Trees or CART, in order to characterize which patients responded to mAb therapy as measured by post-therapy CD137 expression. We supplemented this with sensitivity analyses using two other predictive modeling tools, Least absolute shrinkage and selection operator (Lasso) regression, and Random forest, where the latter predictive approaches address different questions than those addressed by CART. Details of these tools are provided in Supplementary Methods, available at Annals of Oncology online.
Finally, we examined the difference (post–pre mAb therapy) in CD137 expression by cancer type using the Wilcoxon matched-pairs signed rank test. For the latter, testing was two-sided and conducted at the 0.05 level of significance. Analyses were conducted using SAS Version 9.4 (Cary, NC), R [15–17], and Graphpad Prism.
Results
A total of 199 eligible patients were enrolled. The demographics and clinicopathologic characteristics of the study population by cancer type are shown in supplementary Table S1, available at Annals of Oncology online. 62 (31%) patients had breast cancer (BC), 46 (23%) had head and neck cancer (HNC) and 91 (46%) had non-Hodgkin’s lymphoma (NHL). Most patients (68%) were <70 years old. For BC and NHL, the majority of patients had FcγRIIIA-158 polymorphism with low affinity alleles (F/F), 57% and 63%, respectively. About half the patients with HNC had FcγRIIIA-158 polymorphism with low affinity alleles (F/F). Most patients (60%) had less than two prior lines of therapy.
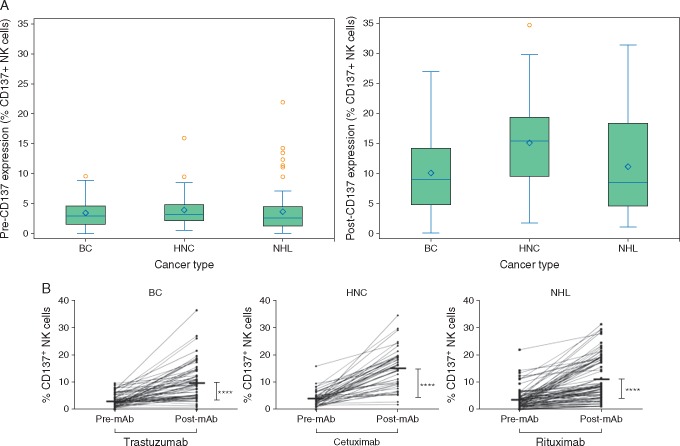
PBMCs were isolated prior to and following the first mAb administration; CD137 was examined on NK cells by flow cytometry (supplementary Figure S1, available at Annals of Oncology online and Figure 1). In support of our previous findings in smaller cohorts of patients [10–12], post-mAb CD137 expression increased significantly across all patients and for each cancer type (Figure 1B; P < 0.001, supplementary Table S2, available at Annals of Oncology online). The difference in post-mAb CD137 from baseline was heterogenous, and was highest for patients with HNC [Median (IQR): 11.5 (5.6–14.3)].
Figure 1.
CD137 increases on circulating NK cells in patients receiving mAb therapy. PBMCs were isolated from 199 patients with breast cancer (BC), head and neck cancer (HNC), and Non Hodgkin lymphoma (NHL). (A) Distribution of pre-mAb and post-mAb CD137 expression (% CD137+ NK cells) as a box plot for each cancer type. The left panel shows pre-mAb CD137 expression and the right panel shows post-mAb CD137 expression. The horizontal line in each box shows the median value and the diamond shows the mean value. (B) Percentage of CD137+ cells among circulating CD3–CD56+ NK cells from 199 patients prior to and following mAb therapy, stratified by tumor type (mean at each time point denoted by bar). ****P ≤ 0.0001.
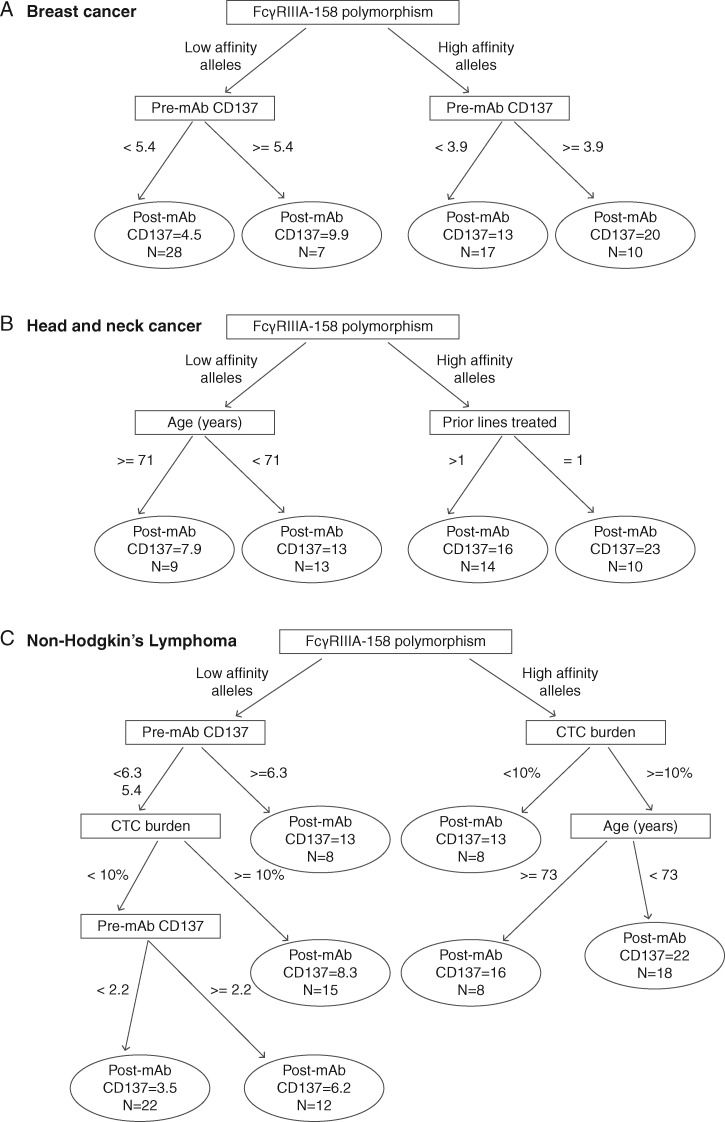
For BC, CART found FcγRIIIA-158 polymorphism to be the most important predictor of post-mAb CD137 expression, followed by pre-mAb CD137. High post-mAb CD137 expression was predicted among patients with high affinity alleles for FcγRIIIA-158 polymorphism who had pre-mAb CD137 expression ≥ 3.9 (Figure 2A). The Random forest results similarly found FcγRIIIA-158 polymorphism to be the most important predictor of post-mAb CD137 but did not attach as much importance to pre-mAb CD137. Not including FcγRIIIA-158 polymorphism in the predictive model would decrease model accuracy by 25%; not including pre-mAb CD137 would decrease model accuracy by 5% (supplementary Figure S2A, available at Annals of Oncology online). Lasso found similar results to CART where FcγRIIIA-158 polymorphism was selected as the most important predictor. Additionally, two other predictors were selected: (i) an interaction between pre-mAb CD137 expression and FcγRIIIA-158 polymorphism, suggesting those with higher pre-mAb CD137 expression and the high affinity alleles had increased post-mAb CD137 relative to those with low affinity alleles; and (ii) an interaction between FcγRIIIA-158 polymorphism (low affinity alleles) and HER2 level indicating those with the polymorphism and low HER2 levels had decreased post-mAb CD137 relative to those without the polymorphism (supplementary Table S3, available at Annals of Oncology online).
Figure 2.
CART prediction results by cancer type. (A) Breast cancer. (B) Head–neck cancer. (C) Non-Hodgkins lymphoma.
For HNC, CART found FcγRIIIA-158 polymorphism to be the most important predictor of post-mAb CD137 expression, followed by age and prior lines of treatment. High post-mAb CD137 expression was predicted among patients with high affinity alleles for FcγRIIIA-158 polymorphism who had one prior treatment line (Figure 2B). The Random forest results similarly found FcγRIIIA-158 polymorphism to be the most important predictor of post-mAb CD137; not including FcγRIIIA-158 polymorphism in the predictive model would decrease model accuracy by ∼21%. In contrast, age and prior lines of treatment had less importance; not including them would decrease model accuracy 2% and 4%, respectively (supplementary Figure S2B, available at Annals of Oncology online). Lasso found similar results to CART; two interaction terms that predicted post-mAb CD137 were selected: (i) interaction between age and FcγRIIIA-158 polymorphism, which predicted that low affinity alleles and older age had lower post-mAb CD137 expression relative to high affinity alleles and lower age, and (ii) interaction between FcγRIIIA-158 polymorphism and one prior line of therapy, which predicted that high affinity alleles and prior treatment had higher post-mAb CD137 relative to those with low affinity alleles (supplementary Table S3, available at Annals of Oncology online).
For NHL, CART found FcγRIIIA-158 polymorphism to be the most important predictor of post-mAb CD137 expression (Figure 2C). Circulating tumor cell (CTC) burden and pre-mAb CD137 expression were the next important variables. High post-mAb CD137 was predicted among patients with high affinity alleles who had a high (≥ 10%) CTC burden and who were <73 years of age (Figure 2C). Similar results were found by Random forest: not including FcγRIIIA-158 polymorphism in the predictive model would decrease model accuracy by ∼32%. Not including CTC burden and pre-mAb CD137 would decrease accuracy by 19% and 15%, respectively (supplementary Figure S2C, available at Annals of Oncology online). Lasso found different results from CART and Random forest; no independent variables were selected as predictors of post-mAb CD137.
All three approaches were consistent in finding FcγRIIIA-158 polymorphism as the most important predictor of post-mAb CD137 expression. Results from CART differed slightly for BC compared to Lasso and Random forest for other variables; CART found pre-mAb CD137 to be the next important predictor, Lasso found a higher estimated coefficient for prior lines treated, and Random forest did not find either of these variables to be important predictors. For HNC, all three approaches yielded similar results; however, Random forest found prior lines treated and age to be far less important than FcγRIIIA-158 polymorphism in predicting post-mAb CD137. For NHL, CART and Random forest had similar findings but Lasso results differed.
Discussion
A key challenge to mAb therapies has been to identify predictors of response. Although tumor expression of the targeted antigen is necessary for mAb efficacy, its use as a biomarker correlates poorly with response to therapy. For example, high HER2 expression in BC does not predict response to trastuzumab since HER2 can be downregulated as an acquired mechanism of resistance [18, 19]. Similarly, EGFR expression in colorectal and head and neck cancer is not predictive of response to cetuximab, and other biomarkers (KRAS and BRAF V600E mutations) are still not clearly predictive, likely due to heterogeneity among patients and within a patient’s tumor developing during therapy via selection pressures [20, 21]. Given the complex interplay between tumor cells and immune cells, there is an unmet need for robust predictive biomarkers that reflect the dynamic nature of the immune response induced by mAbs, which should be examined not only before therapy start but also during the treatment process.
Clinical results have shown that patients harboring the higher affinity FcγRIIIA polymorphism have better responses to trastuzumab, cetuximab and rituximab [22–24]. In support of these observations, we have previously shown that NK cells from healthy donors with high-affinity FcγRIIIA-158 polymorphisms (V/V or F/V) expressed increased levels of CD137 compared with NK cells from donors with the low affinity (F/F) polymorphism post-exposure to trastuzumab-coated BC cells in vitro [11]. In a small cohort of HNC patients receiving cetuximab, a greater increase in CD137 expression was seen among patients harboring high-affinity alleles FcγRIIIA-158 compared with those with low-affinity alleles [12]. Our current findings confirm these observations and highlight the importance of NK-mediated ADCC in mAb therapies.
The second strongest predictor of post-mAb CD137 expression differed by cancer type. For BC, higher pre-mAb CD137 expression levels predicted higher post-mAb CD137 expression levels, suggesting that the basal activation status of NK cells influences subsequent activation by tumor-targeting mAbs. We also examined the influence of prior exposure to trastuzumab, which has been shown to worsen clinical outcomes for metastatic breast cancer patients [25], and found it not to be predictive of CD137 upregulation, as was the expression level of HER2. This suggests that minimal expression of HER2 by tumors that allows the binding of trastuzumab can sufficiently activate NK cells and upregulate CD137.
For HNC, the HPV status was not predictive of post-mAb CD137 expression levels. Recent retrospective analyses have shown that cetuximab has similar benefit in HPV-positive and HPV-negative patients [26, 27]. In support of this, it has been shown that HNC patients with not only high but also low EGFR-expressing tumors can respond to cetuximab. As in BC, our finding that CD137 upregulation is not influenced by the HPV status of HNC (or, by extension, the EGFR expression level) suggests that minimal binding of cetuximab to EGFR can sufficiently activate NK cells and upregulate CD137.
For NHL, higher pre-mAb CD137 expression levels and higher CTC tumor burden (≥ 10%) were predictive of higher CD137 upregulation on NK cells post-mAb therapy. These results support our previous findings of the correlation between the degree of CD137 upregulation and percent CTCs [10], and suggest that CD137 upregulation on NK cells in NHL is influenced by effector:target ratios, where the target is CTCs bound by rituximab. Another possibility is the effect of CD20 antigenic load on CTC [28]. Interestingly, our results predict that a higher CTC burden in patients carrying the high affinity FcγRIIIa-158 alleles results in higher CD137 upregulation than in patients carrying the low-affinity allele. Dall’Ozzo et al. showed that in vitro, NK cells carrying the high-affinity FcγRIIIa-158 allele require much lower rituximab concentrations to efficiently induce ADCC than NK cells carrying the low-affinity alleles. These functional differences were seen at low rituximab concentrations weakly sensitizing CD20 (resulting in 50% lysis; EC50) but were lost at high rituximab concentrations [29]. Given the inverse correlation between rituximab serum level and both tumor burden and lymphocyte count at baseline [30], this may explain why high CTC counts (or low rituximab concentrations) preferentially upregulate CD137 in high-affinity versus low-affinity NK cells.
Our current study confirms previous findings of heterogeneity in CD137 upregulation [10–12], and identifies several clinical and pathologic characteristics that predict post-mAb CD137 expression. The results highlight the role of FcγRIIIA-158 polymorphism and pre-mAb CD137 expression levels in mAb-induced NK cell activation in all three cancer types, in addition to age (for HNC and NHL), number of prior-line therapies (for HNC), and CTC burden (for NHL). A limitation of our current study is that we did not validate our findings in an independent cohort; however, the consistency of our findings across the three approaches suggests that our results are reliable. Of note, the clinical outcome data for the current trial was not available to address whether CD137 upregulation on NK cells correlates with objective responses and/or progression-free survival (PFS) in patients receiving mAb therapies.
Our ultimate goal is to build a nomogram predictive of CD137 upregulation in patients receiving mAb therapies. Towards that goal, the next step is to validate our predictive model using additional data to obtain metrics (e.g. area under the curve, AUC) that assess the predictive accuracy of the model. Depending on these findings, the following step would be to build the nomogram and to validate it using large multi-institutional datasets of patients receiving mAb therapies from ongoing clinical trials.
The nomogram can aid clinicians in more accurate decision-making for patient selection not only for standard mAb therapies but also for investigational trial design of combination therapies with anti-CD137. These decisions can be further supported with in vitro studies using anti-CD137-activated patient NK cells and ex vivo patient tumor samples, which functionally assess the increased CD137 expression on NK cells as measured by increased NK anti-tumor cytotoxicity.
As promising new tumor-targeting mAb therapies are developed, the need to accurately identify patient populations most likely to benefit from them becomes increasingly important. Nomograms capture the inherent variation in individual responses to therapies and, when designed based on highly relevant biological markers, hold the promise of improving prediction and patient selection, maximizing therapy efficacy, and reducing toxicity and costs.
Supplementary Material
Acknowledgements
We wish to thank the patients and their families. We also thank members of the clinical trial team and laboratory staff at the Stanford Cancer Institute who have contributed to this work. This article is dedicated to the memory of H.K., who pushed to make a difference in this world where time is always too short, so that the future is different for cancer patients, and for the better.
Funding
This work was supported by funding from the Leukemia and Lymphoma Society (Career Development Program—Scholar in Clinical Research 2016) and from the AntiCancer Fund (no applicable grant number).
Disclosure
The authors have declared no conflicts of interest.
References
- 1. Geynisman DM, Chien CR, Smieliauskas F. et al. Economic evaluation of therapeutic cancer vaccines and immunotherapy: a systematic review. Hum Vaccin Immunother 2014; 10: 3415–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scolnik PA. mAbs: a business perspective. MAbs 2009; 1: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartkowiak T, Curran MA.. 4-1BB agonists: multi-potent potentiators of tumor immunity. Front Oncol 2015; 5: 117.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ascierto PA, Simeone E, Sznol M. et al. Clinical experiences with anti-CD137 and anti-PD1 therapeutic antibodies. Semin Oncol 2010; 37: 508–516. [DOI] [PubMed] [Google Scholar]
- 5. Kattan MW. Nomograms. Introduction. Semin Urol Oncol 2002; 20: 79–81. [PubMed] [Google Scholar]
- 6. Kattan MW, Eastham JA, Stapleton AM. et al. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst 1998; 90: 766–771. [DOI] [PubMed] [Google Scholar]
- 7. Shariat SF, Karakiewicz PI, Godoy G, Lerner SP.. Use of nomograms for predictions of outcome in patients with advanced bladder cancer. Ther Adv Urol 2009; 1: 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shariat SF, Capitanio U, Jeldres C, Karakiewicz PI.. Can nomograms be superior to other prediction tools? BJU Int 2009; 103: 492–495. discussion 495–497. [DOI] [PubMed] [Google Scholar]
- 9. Shariat SF, Karakiewicz PI, Suardi N, Kattan MW.. Comparison of nomograms with other methods for predicting outcomes in prostate cancer: a critical analysis of the literature. Clin Cancer Res 2008; 14: 4400–4407. [DOI] [PubMed] [Google Scholar]
- 10. Kohrt HE, Houot R, Goldstein MJ. et al. CD137 stimulation enhances the antilymphoma activity of anti-CD20 antibodies. Blood 2011; 117: 2423–2432. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Kohrt HE, Houot R, Weiskopf K. et al. Stimulation of natural killer cells with a CD137-specific antibody enhances trastuzumab efficacy in xenotransplant models of breast cancer. J Clin Invest 2012; 122: 1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Kohrt HE, Colevas AD, Houot R. et al. Targeting CD137 enhances the efficacy of cetuximab. J Clin Invest 2014; 124: 2668–2682. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13. Caligiuri MA. Human natural killer cells. Blood 2008; 112: 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koene HR, Kleijer M, Algra J. et al. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood 1997; 90: 1109–1114. [PubMed] [Google Scholar]
- 15. Liaw A, Weiner M. Classification and Regression by randomForest. R News 2002; 2: 18–22.
- 16. Development Core Team R. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing 2014.
- 17. Therneau T, Atkinson B, Ripley B. rpart: recursive partitioning, 4.1-3 edition. R package 2013; http://CRAN.R-project.org/package=rpart (1 December 2017, date last accessed).
- 18. Chung A, Cui X, Audeh W, Giuliano A.. Current status of anti-human epidermal growth factor receptor 2 therapies: predicting and overcoming herceptin resistance. Clin Breast Cancer 2013; 13: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guarneri V, Dieci MV, Barbieri E. et al. Loss of HER2 positivity and prognosis after neoadjuvant therapy in HER2-positive breast cancer patients. Ann Oncol 2013; 24: 2990–2994. [DOI] [PubMed] [Google Scholar]
- 20. Luo HY, Xu RH.. Predictive and prognostic biomarkers with therapeutic targets in advanced colorectal cancer. World J Gastroenterol 2014; 20: 3858–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kosmidou V, Oikonomou E, Vlassi M. et al. Tumor heterogeneity revealed by KRAS, BRAF, and PIK3CA pyrosequencing: KRAS and PIK3CA intratumor mutation profile differences and their therapeutic implications. Hum Mutat 2014; 35: 329–340. [DOI] [PubMed] [Google Scholar]
- 22. Tamura K, Shimizu C, Hojo T. et al. FcγR2A and 3A polymorphisms predict clinical outcome of trastuzumab in both neoadjuvant and metastatic settings in patients with HER2-positive breast cancer. Ann Oncol 2011; 22: 1302–1307. [DOI] [PubMed] [Google Scholar]
- 23. Bibeau F, Lopez-Crapez E, Di Fiore F. et al. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol 2009; 27: 1122–1129. [DOI] [PubMed] [Google Scholar]
- 24. Weng WK, Levy R.. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol 2003; 21: 3940–3947. [DOI] [PubMed] [Google Scholar]
- 25. Murthy RK, Varma A, Mishra P. et al. Effect of adjuvant/neoadjuvant trastuzumab on clinical outcomes in patients with HER2-positive metastatic breast cancer. Cancer 2014; 120: 1932–1938. [DOI] [PubMed] [Google Scholar]
- 26. Psyrri A, Sasaki C, Vassilakopoulou M. et al. Future directions in research, treatment and prevention of HPV-related squamous cell carcinoma of the head and neck. Head Neck Pathol 2012; 6(Suppl 1): S121–S128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samuels SE, Eisbruch A, Beitler JJ. et al. Management of locally advanced HPV-related oropharyngeal squamous cell carcinoma: where are we? Eur Arch Otorhinolaryngol 2015; 10:2877–2894. [DOI] [PubMed] [Google Scholar]
- 28. Cartron G, Trappe RU, Solal-Céligny P, Hallek M.. Interindividual variability of response to rituximab: from biological origins to individualized therapies. Clin Cancer Res 2011; 17: 19–30. [DOI] [PubMed] [Google Scholar]
- 29. Dall’Ozzo S, Tartas S, Paintaud G. et al. Rituximab-dependent cytotoxicity by natural killer cells: influence of FCGR3A polymorphism on the concentration-effect relationship. Cancer Res 2004; 64: 4664–4669. [DOI] [PubMed] [Google Scholar]
- 30. Berinstein NL, Grillo-López AJ, White CA. et al. Association of serum Rituximab (IDEC-C2B8) concentration and anti-tumor response in the treatment of recurrent low-grade or follicular non-Hodgkin’s lymphoma. Ann Oncol 1998; 9: 995–1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.