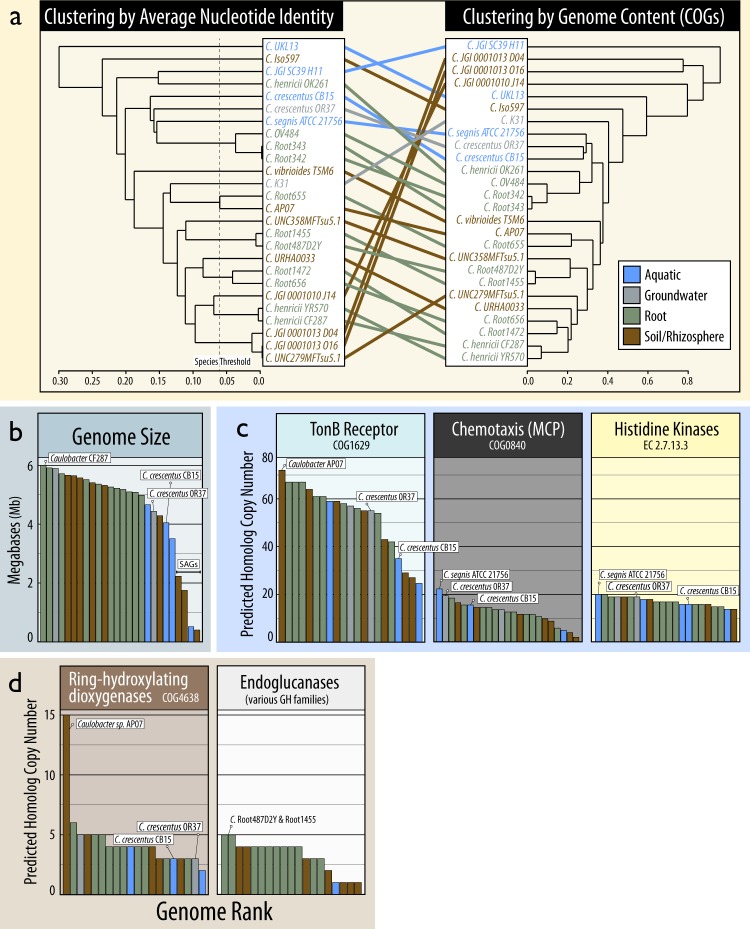
Fig. 6.
Comparisons of Caulobacter genomes based on (a) their phylogenetic and functional relatedness, (b) genome size, (c) abundance of transport, chemotaxis, and signaling genes identified as signatures of oligotrophy in Caulobacter [8], and (d) catabolic genes commonly involved in decomposing lignocellulose. In a the phylogenetic relatedness is based on average nucleotide identity (1 – ANI) and functional relatedness is based on Bray-Curtis dissimilarity of COG profiles. The ANI threshold associated with separate species are indicated (Konstantinidis and Tiedje, 2005). The dendrograms in (a) were ordered using the ladderize function. In (b–d), the rank location of the type strain, C. crescentus CB15, and the closely related C. crescentus OR37 are displayed (when present) and the genome(s) with the highest number of copies of each gene family