FIGURE 1.
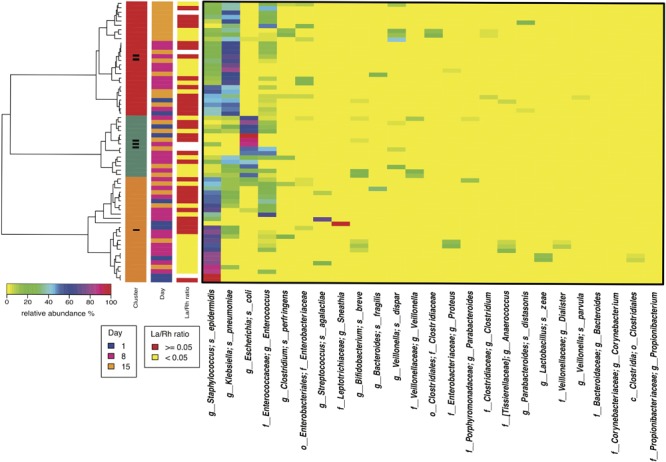
Heatmap of the 50 most abundant intestinal bacterial taxa relative abundance in samples collected from 38 preterm infants enrolled in the study. The microbiota of 64 fecal samples were successfully characterized by high-throughput sequencing of the V3–V4 variable regions of 16S rRNA genes. The three sidebars indicate cluster, time, and intestinal permeability category, respectively. Ward linkage clustering was used to cluster samples based on their Jensen-Shannon distance calculated in vegan package in R (Oksanen et al., 2011). The samples with no IP assessment were included to generated the clusters. The low and high intestinal permeability category was defined by a La/Rh > 0.05 or < = 0.05 respectively (Saleem et al., 2017). Taxonomic profiling of corresponding metagenomes further resolved Klebsiella spp. to Klebsiella pneumoniae, Enterococcus spp. to Enterococcus faecalis, and Bifidobacterium spp. to Bifidobacterium breve.
