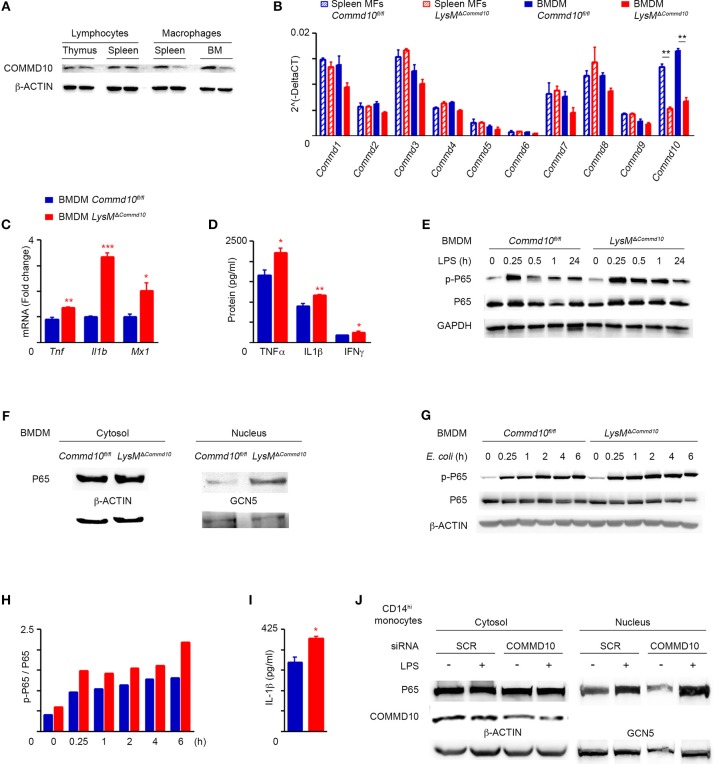
Figure 1.
LysMΔCommd10 BMDM respond to LPS by higher production of pro-inflammatory cytokines. (A,B) Cells were isolated from spleen and BM of Commd10fl/fl (blue) or LysMΔCommd10 (red) mice. (A) Immunoblots showing the expression of COMMD10 in cell lysates of thymic or splenic lymphocytes vs. splenic or BM-derived macrophages. β-actin was utilized as control. (B) qRT-PCR gene expression of all COMMD family members in splenic macrophages and BMDM (n = 3). (C,D) BMDM were stimulated with LPS (100 ng/ml) for 3 h. (C) qRT-PCR gene expression of pro-inflammatory cytokine in cell lysate (n = 3). (D) ELISA of pro-inflammatory cytokine levels in cell free supernatants (n = 3). (E) Representative immunoblot of BMDM cell lysates following LPS challenge (100 ng/ml) showing the protein expression of phospho-P65 (p-P65) and P65 over time course. GAPDH was used as control. (F) Immunoblots showing P65 expression in cytosolic and nuclear fraction lysates of LPS-treated BMDM. β-ACTIN and Polymerase II antibodies, respectively, were used as controls (n = 3). (G–I) BMDM from Commd10fl/fl or LysMΔCommd10 mice were infected with E. Coli at multiplicity of infection (MOI) = 1. (G) Immunoblots showing the protein expression of p-P65 and P65 over time course. β-ACTIN was used as control. (H) Respective densitometry-based quantification of p-P65/P65 ratio in Commd10fl/fl vs. LysMΔCommd10 BMDM over time course following E. coli infection. (I) ELISA analysis of IL-1β from supernatants at 6 h post-infection with E. coli (n = 4). (J) Immunoblots showing cytosolic COMMD10 protein expression following siRNA-based gene silencing vs. scrambled siRNA control, β-ACTIN was used as control, and immunoblots showing P65 expression in cytosolic and nuclear fraction lysates of LPS-treated human CD14hi monocyte-derived macrophages. GCN5 was used as control in the nuclear fraction (n = 3). Data were analyzed by unpaired, two-tailed t-test, comparing each time between Commd10fl/fl and LysMΔCommd10 (red stars), and are presented as mean ± SEM with significance: *p < 0.05, **p < 0.01, ***p < 0.001. Data in (A,B,E) represents two-independent experiments. Data in (C,D,F–J) are from a single experiment.