Host phagocytic cells are crucial players in initial defense against Candida albicans infection. C. albicans utilizes MAP kinases and Ras1 stress response signaling pathways to protect itself from killing by immune cells.
KEYWORDS: Candida albicans, human neutrophils, phagocytosis, Ras1
ABSTRACT
Host phagocytic cells are crucial players in initial defense against Candida albicans infection. C. albicans utilizes MAP kinases and Ras1 stress response signaling pathways to protect itself from killing by immune cells. In this study, we tested the importance of these pathways in C. albicans phagocytosis by neutrophils and subsequent phagosomal survival. Phagocytosis was influenced by C. albicans morphology, so hyphal length of >10 μm reduced the phagocytic index (PI) 2- to 3-fold in human neutrophils. Primary human neutrophils killed 81% of phagocytosed C. albicans, while primary mouse neutrophils killed 63% of yeasts. We found that both the C. albicans Cek1 and Hog1 pathways were required for survival of phagocytosed yeast, whereas deletion of C. albicans RAS1 resulted in an 84% increase in survival within neutrophils compared to that of the wild type (WT). The absence of Ras1 did not alter reactive oxygen species (ROS) production by C. albicans; however, phagocytosed C. albicans Δ/Δras1 cells reduced ROS release by neutrophils by 86%. Moreover, C. albicans Δ/Δras1 cells had increased resistance to hydrogen peroxide as a result of high levels of catalase activity. This phenotype was specific to Ras1, since these effects were not observed in the absence of its partner Cyr1 or with its downstream target Efg1. In addition, C. albicans Δ/Δras1 cells had a significantly increased resistance to nonoxidative killing by human neutrophil peptide 1 (HNP-1) that was reversed by restoring cellular cAMP levels. These data show that C. albicans Ras1 inactivation leads to fungal resistance to both oxidative and nonoxidative mechanisms of neutrophil phagosomal killing.
INTRODUCTION
Candida albicans is an opportunistic fungus colonizing the oral cavity in over half of healthy individuals (1); however, when the immune system is compromised, this normally commensal organism may become pathogenic and cause oropharyngeal candidiasis (OPC). Recruitment of phagocytic cells is considered a crucial early response to mucosal invasion by C. albicans. In a murine model of OPC, neutrophils migrate to the superficial epithelium and are essential for clearance of fungi within 1 day of infection (2). Humans and mice with innate immune deficiencies of neutrophils or macrophages have an elevated risk of invasive candidiasis and increased mortality rates (3, 4).
Professional phagocytes, including neutrophils and macrophages, function in engagement and phagocytosis of invading microbes, although their relative importance in killing and clearance of C. albicans differs. Following recognition of C. albicans, the two cell types engulf yeast through the process of phagocytosis at similar rates (5). However, once phagocytosed, neutrophils are more efficient at killing C. albicans than macrophages (80% and 60% killing rates, respectively) (6), and human neutrophils have significantly higher C. albicans killing activity than murine neutrophils (7). The high efficiency of human neutrophil killing of C. albicans is likely due to production of small reactive oxygen species (ROS) as well as human neutrophil peptide 1 (HNP-1), although the relative contributions of oxidative and nonoxidative mechanisms are unknown.
Killing of engulfed microbes within neutrophil phagosomes is initiated with a powerful oxidative burst produced by the NADPH-oxidase complex (NOX2) that is assembled on the membrane of the maturing phagosome. The NOX2 complex forms superoxide in the lumen of the phagosome, which goes on to form other small ROS and reactive nitrogen species (RNS), including H2O2, NO2•, and OCl−. These compounds are highly concentrated within the phagosomal lumen so that ROS and RNS are efficient at killing fungi engulfed within this space (reviewed in reference 8). Despite the similarities between human and mouse neutrophil ROS release, production of ROS by human granulocytes is significantly higher than their mouse counterpart driven by a higher myeloperoxidase activity, which relates to the higher killing activity (7). In addition, human neutrophil phagosomes maintain NOX2 activity for an extended period (9) and are capable of delivering their preformed granules containing antimicrobial components by fusion with the maturing phagosome. HNP-1, commonly known as alpha-defensin, is one of the most abundant cationic peptides contained in azurophilic granules of human neutrophils. The presence of HNP-1 makes human neutrophils highly efficient in controlling C. albicans compared to mouse neutrophils, which lack alpha-defensins (7).
Although phagocytosis and subsequent killing are highly efficient and controlled processes, C. albicans and other pathogenic microbes are often able to adapt and resist these immune defenses in order to survive encounters with neutrophils (10–12). For example, C. albicans activates signal transduction pathways, particularly mitogen-activated protein kinase (MAP kinase) and Ras1-GTPase, which are crucial for C. albicans sensing of environmental conditions and initiating survival responses. C. albicans has three major MAP kinase signal transduction pathways involved in cellular and biological processes that have been widely studied (reviewed in references 13 and 14). The Cek1 pathway is a crucial contributor to cell wall biogenesis, mating, and morphogenesis (15–17). The C. albicans Hog1 pathway responds to osmotic and oxidative conditions, with Hog1 deletion mutants being more susceptible to hydrogen peroxide exposure (18) as well as being hyperfilamentous (18). The third MAP kinase is Mkc1, which functions during cell wall biogenesis and invasive growth and has an important role during biofilm formation (17).
In addition to the three MAP kinase pathways, the Ras1-cAMP pathway is also linked to morphogenesis and responses of C. albicans to environmental stress. This pathway is composed of the small Ras1-GTPase protein, the adenylyl cyclase Cyr1, and the downstream transcription factor Efg1. Although Cyr1 was originally thought to be a downstream component of the Ras1-cAMP pathway and fully dependent upon Ras1 activity, recent studies indicate a parallel and complex model in which the two components regulate each other (19, 20). Cyr1 senses high intracellular levels of ATP and triggers the exchange of Ras1-bound GDP for GTP, causing Ras1 activation (19). After Cyr1/Ras1 activation, ATP is converted to cAMP, which derepresses protein kinase A (PKA), leading to the activation of Efg1. Efg1, in turn, is important for filamentation and other biological processes (reviewed in reference 21). A variety of extracellular stimuli are capable of activating the Ras1 pathway, including low pH and serum. Furthermore, both Ras1 and Cyr1 are needed for virulence in a disseminated model of murine candidiasis (22, 23).
The connections between the MAP kinase signaling pathways and innate immunity have been broadly studied, and each pathway has been found to contribute to the initiation and maintenance of systemic infection in murine candidiasis (16, 24, 25). Galán-Díez et al. demonstrated that in the absence of CEK1, C. albicans has higher exposed β-glucan, which correlates with increased phagocytosis by human macrophages (26). Also, deletion of selected regulator components of the C. albicans Hog1 pathway results in increased susceptibility to human neutrophil killing (27). However, nothing is currently known about the relationship between C. albicans Ras1 and its interplay with immune cells, other than evidence from a recent transcriptomic study of C. albicans phagocytosed by human neutrophils indicating that certain Ras1 pathway components were downregulated (28), suggesting that dampening of Ras1 signaling is important for survival of phagocytosed C. albicans.
In addition to being important regulators of the Ras1 pathway, levels of cAMP and ATP also affect major yeast virulence attributes. Increased intracellular levels of cAMP trigger C. albicans hyperfilamentation (29), while reduced cAMP initiates increased expression of genes required for stress defenses, leading to increased survival (30, 31). Additionally, low intracellular ATP levels in C. albicans lead to altered cell wall composition and physical state of the membrane and are associated with increased resistance to cationic antifungal peptides (32). Thus, it is likely that the Ras1 pathway is at least partially linked to these known cAMP and ATP effects on virulence and plays an important role in survival of C. albicans organisms that have been internalized by neutrophils.
In this study, we investigated the role of the fungal MAP kinases and Ras1 pathway in phagocytosis and intracellular killing of yeast by human neutrophils. We found that both Hog1 and Cek1 MAP kinases were required for survival of C. albicans phagocytosed by primary human neutrophils. However, C. albicans RAS1 knockouts had significantly increased survival within neutrophils compared to wild-type (WT) cells. This increased survival of C. albicans RAS1 knockouts was due, at least partially, to their increased resistance to both oxidative stress and HNP-1 as well as inhibition of ROS production by human neutrophils. Therefore, the Ras1 pathway plays a crucial role during C. albicans defense against killing by neutrophils.
RESULTS
Efficiency of phagocytosis of C. albicans is determined by hyphal length.
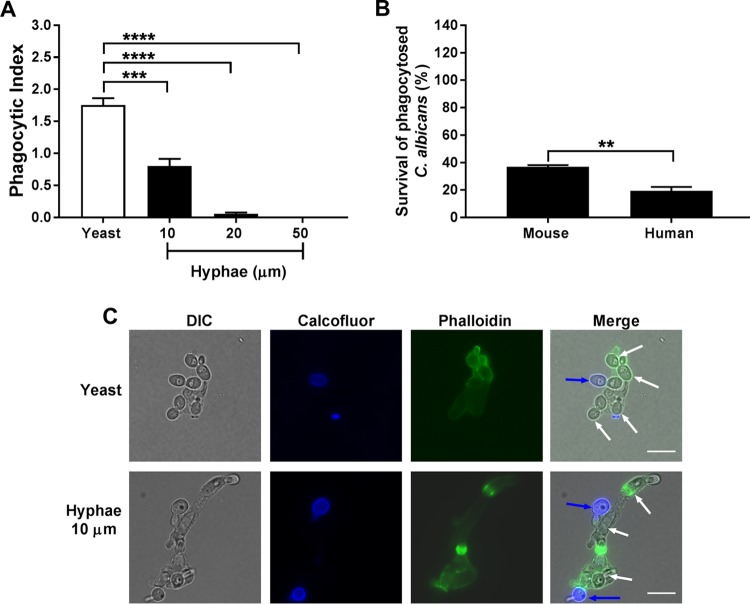
Since phagocytosis is dependent upon the morphology of the target cell (33), we first examined the effect of C. albicans hyphal length on phagocytosis by human blood-derived neutrophils (H-PMNs) after 30 min of incubation at a multiplicity of infection (MOI) of 3. The phagocytic index (PI) of C. albicans with preformed hyphae was significantly reduced compared to that of yeast form cells (Fig. 1A). The PI of C. albicans with an average hyphal length of 10 μm was reduced 2-fold and increasing hyphal length further reduced the PI, so neutrophils had little to no success at phagocytosis of C. albicans cells with hyphae with an average length of 50 μm (Fig. 1A and C). Yeast cells were efficiently engulfed by H-PMNs and were frequently observed to contain 3 or 4 yeast cells (Fig. 1C). H-PMNs showed remarkable plasticity in engulfment of C. albicans hyphal cells with a length of 10 μm, so the overall dimensions of the neutrophil were increased almost two times the original size during phagocytosis of two hyphal cells (Fig. 1C). As we found that hyphal formation reduced the PI, for further experiments only yeast form cells of C. albicans were used, although some mutant strains had a hyperfilamentous phenotype, so some hyphae were formed. Next, we assessed the relative abilities of primary mouse and human neutrophils to kill internalized phagocytosed C. albicans after 2.5 h. We found that H-PMNs had a higher rate of killing (only 19% of C. albicans yeast survived) than did primary murine-bone marrow derived neutrophils (M-PMNs), in which 37% of internalized C. albicans organisms survived (Fig. 1B). Therefore, H-PMNs were used for all subsequent experiments based on this high killing rate for C. albicans.
FIG 1.
Initial assessment of C. albicans phagocytosis and survival by neutrophils. (A) Human blood-derived neutrophils (H-PMNs) were infected with C. albicans yeast or 10-, 20-, or 50-μm hyphae at an MOI of 3 for 30 min. Extracellular fungal cells were stained using calcofluor, and H-PMNs were stained with phalloidin. Unstained intracellular C. albicans cells were counted in at least 100 H-PMNs, and the phagocytic index was calculated by obtaining the ratio of the total number of ingested C. albicans cells and the total number of neutrophils counted. (B) Murine or human primary neutrophils were coincubated with C. albicans at an MOI of 0.1 for 3 h. Cells were lysed and internalized C. albicans was released, plated in agar, and incubated for 48 h to obtain viable CFU. Survival was calculated as follows: (recovered C. albicans CFU after neutrophil lysis/total number of phagocytosed C. albicans cells) × 100. Results are the means ± SEs from at least three independent experiments performed in duplicate. Significance was calculated using one-way ANOVA with post hoc Dunnett’s multiple-comparison test (A) or unpaired Student's t test (B). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (C) Representative yeast and hyphal (average of 10 μm) phagocytosis by H-PMNs. Nonphagocytosed C. albicans cells are shown by calcofluor white staining (blue arrows), H-PMNs by phalloidin staining (green), and phagocytosed C. albicans by white arrows in merged panels. Scale bars = 10 μm.
MAP kinases and Ras1 are important contributors to C. albicans survival within human neutrophils.
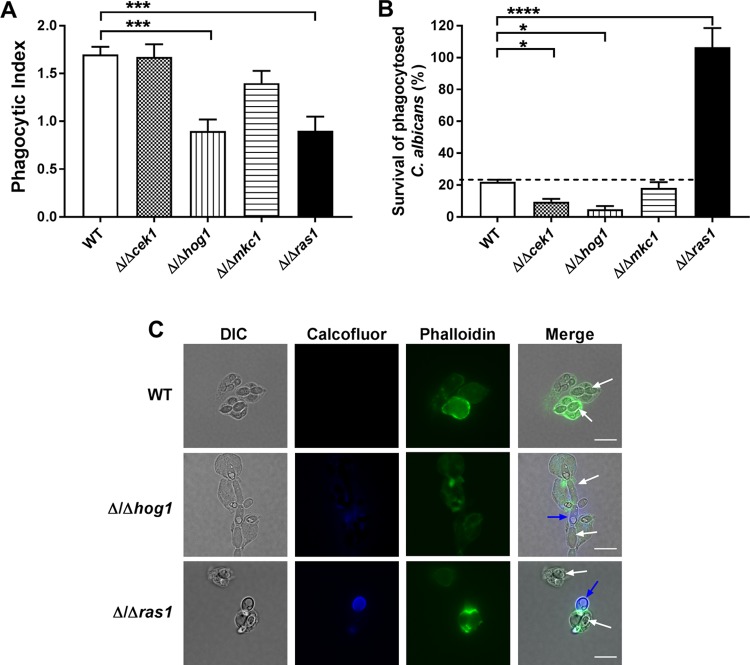
C. albicans MAP kinase signaling pathways are involved in resistance to oxidative and environmental stresses; however, little is known about any role these pathways play in modulating survival within professional phagocytic cells. Therefore, C. albicans mutants in which the major stress response signaling pathways had been knocked out (Δ/Δcek1, Δ/Δhog1, Δ/Δmkc1 and Δ/Δras1) were tested for their ability to be phagocytosed and survive within H-PMNs. Significant differences (P < 0.001, one-way analysis of variance [ANOVA]) were found in the PIs of C. albicans HOG1 and RAS1 strains and WT cells, while no differences in phagocytosis were measured in MKC1 or CEK1 knockouts (Fig. 2A). The PI of C. albicans Δ/Δhog1 and Δ/Δras1 mutants was reduced by 50% compared to that of the WT. Surprisingly, C. albicans Δ/Δcek1 cells with defects in cell wall composition and thus expected to have defects in neutrophil recognition and uptake had no difference in phagocytosis compared to that of the WT (Fig. 2A).
FIG 2.
C. albicans CEK1 and HOG1 deletion mutants have decreased survival within human neutrophils, while RAS1 deletion mutants have increased survival. (A) H-PMNs were infected with WT, Δ/Δcek1, Δ/Δhog1, Δ/Δmkc1, and Δ/Δras1 C. albicans strains at an MOI of 3 for 30 min. Extracellular fungal cells were stained using calcofluor white, and H-PMNs were stained with phalloidin. Unstained intracellular C. albicans cells were counted in at least 100 phagocytic cells, and the phagocytic index was calculated by obtaining the ratio of the total number of ingested C. albicans and the total number of phagocytes counted. (B) To evaluate survival, H-PMNs were infected with the same C. albicans strains in an MOI of 0.1 for 3 h. After coincubation, phagocytic cells were lysed and internal C. albicans was released, plated in agar, and incubated for 48 h to obtain viable CFU. Survival was calculated as follows: (recovered C. albicans CFU after neutrophil lysis/total number of phagocytosed C. albicans cells) × 100. Results represent the means ± SEs from at least three independent experiments carried out in duplicate. Significance was obtained using one-way ANOVA with post hoc Dunnett’s multiple-comparison test. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (C) Representative phagocytosis of C. albicans WT, Δ/Δhog1, and Δ/Δras1 after 30 min of infection with H-PMNs. Nonphagocytosed C. albicans cells are shown by calcofluor white staining (blue arrows), H-PMNs by phalloidin staining (green), and phagocytosed C. albicans by white arrows in merged panels. Scale bars = 10 μm.
Differences in the survival of C. albicans MAP kinase knockouts of phagocytosed yeast cells (Fig. 2B) were calculated independently of their PI (see Materials and Methods). As expected, survival of C. albicans Δ/Δcek1 and Δ/Δhog1 mutants (with knockouts of signaling pathways important for response to cell wall and oxidative/osmotic stressors, respectively) was significantly decreased in H-PMNs (13% and 6% survival, respectively) compared to that of WT cells. No difference in survival was found for C. albicans Δ/Δmkc1 within H-PMNs. Unexpectedly, we observed dramatically increased survival of C. albicans Δ/Δras1 cells (106%) that was not observed for any other MAP kinase mutant. These differences were not related to the production of hyphae within phagosomes, since hyperfilamentous C. albicans Δ/Δhog1 cells produced hyphae after phagocytosis (Fig. 2C) but were killed more efficiently that WT cells, while C. albicans Δ/Δras1 cells remained as the yeast form (Fig. 2C) yet were highly resistant to killing.
C. albicans Ras1 is important for resistance to phagosomal killing by human neutrophils.
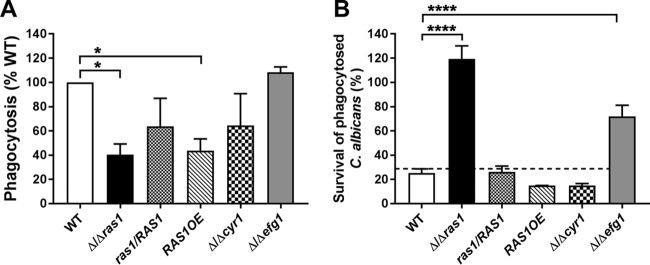
We next examined whether other C. albicans Ras1 pathway components, including its interacting partner Cyr1, or downstream signaling components (Efg1) were needed for resistance to H-PMN killing. To further evaluate the specificity of Ras1 for these phenotypes, a Ras1 overexpression strain (RAS1OE) and complemented ras1/RAS1 strain also were examined. Inhibited phagocytosis of C. albicans Δ/Δras1 cells by H-PMNs was reversed in the complemented ras1/RAS1 strain, while a small but significant defect in phagocytic uptake of RAS1OE cells was retained (Fig. 3A). However, the PIs of Δ/Δcyr1 and Δ/Δefg1 deletion mutants did not differ. Thus, altering intracellular levels of Ras1 likely resulted in changes in the cell surface that affected phagocytic recognition, while Cyr1 or Efg1 did not influence these interactions.
FIG 3.
RAS1 and EFG1 deletions improve survival of C. albicans within human neutrophils. (A) H-PMNs were infected with C. albicans WT, Δ/Δras1, ras1/RAS1, RAS1OE, Δ/Δcyr1, and Δ/Δefg1 strains for 30 min. Extracellular fungal cells were stained using calcofluor white, unstained Candida cells were counted, and the PI was calculated by obtaining the ratio of the total number of ingested C. albicans cells and the total number of phagocytes counted. Phagocytosis of the WT was set as 100%. (B) Survival of C. albicans strains within H-PMNs was evaluated after 3 h of coincubation. H-PMNs were lysed, and internalized C. albicans cells were released, plated in agar, and incubated for 48 h to obtain viable CFU. Survival was calculated as follows: (recovered C. albicans CFU after neutrophil lysis/total number of phagocytosed C. albicans cells) × 100. Results represent the means ± SEs from at least three independent experiments carried out in duplicate. Significance was calculated using one-way ANOVA with post hoc Dunnett’s multiple-comparison test. *, P < 0.05; ****, P < 0.0001.
Next, we compared the high resistance to neutrophil killing of C. albicans Δ/Δras1 cells with the resistance of other strains (Fig. 3B). As expected, complementation of RAS1 (ras1/RAS1) restored neutrophil killing to WT levels, while C. albicans Δ/Δcyr1 and RAS1OE cells showed no significant difference in survival compared to that of WT cells (survival rates of 15% and 14.8%, respectively). On the other hand, C. albicans Δ/Δefg1 cells had increased survival within H-PMNs (72%, compared to 25% for WT); however, this was approximately half the survival rate of C. albicans Δ/Δras1 cells, suggesting that the mechanism of survival within phagosomes contributed by Efg1 differs from that of Ras1.
C. albicans Δ/Δras1 cells suppress ROS production in human neutrophils.
Shortly after completion of phagosome formation, a powerful oxidative burst comprised of reactive oxygen species (ROS) and reactive nitrogen species (RNS) is released into the lumen of the phagosome (34). Therefore, we next measured whether C. albicans Δ/Δras1 cells were able to suppress ROS production within neutrophils. C. albicans Δ/Δras1 cells were compared to WT, complemented, and overexpression strains for the ability to stimulate H-PMN ROS generation using a luminol-based chemiluminescence assay (Fig. 4A). Since C. albicans cells themselves are able to release ROS to the extracellular environment in response to some stimuli (35), ROS production by C. albicans strains alone was measured under the same conditions (Fig. 4B) and these values were subtracted from the total values for ROS generated by neutrophils stimulated with yeasts. Neutrophils stimulated with WT yeast produced ROS by 10 min and had maximal ROS production by 50 min, which was temporally similar to ROS production by the positive-control phorbol myristate acetate (PMA)-stimulated neutrophils that produced almost twice the levels of ROS (in agreement with previous reports [36, 37]). Surprisingly, H-PMNs stimulated with C. albicans Δ/Δras1 had substantially delayed detectable ROS production (no ROS was evident until after 40 min), and levels of ROS were reduced by an average of 86% of that of the WT (Fig. 4A and C). Moreover, detectable ROS production remained suppressed by neutrophils when neutrophils were incubated with C. albicans Δ/Δras1 through 150 min of experimental observation. This effect on ROS production was specific to the Ras1 knockout strain, as the complemented strain and C. albicans Δ/Δefg1 (Fig. 4A, blue line and gray line, respectively) elicited ROS production similar to that of the WT. Furthermore, stimulation of H-PMNs with the RAS1OE overexpression strain resulted in a nearly immediate and large burst of ROS production (Fig. 4A, red dotted line), suggesting that Ras1 overexpression elicits changes in the cell that trigger immediate neutrophil activation and ROS production. This suppression of neutrophil ROS production by C. albicans Δ/Δras1 cells was not due to any differences in ROS production by the yeasts themselves, as all strains except for the RAS1 overexpression strain had similar levels of ROS production (Fig. 4B). Quantification of total ROS production in four independent experiments confirmed that H-PMNs produced significantly less detectable ROS when stimulated with C. albicans Δ/Δras1, while Δ/Δefg1 and RAS1OE cells produced total ROS levels similar to those of the WT (Fig. 4C), suggesting that intraphagosomal survival of Δ/Δras1 cells is related to suppression of ROS production, while Δ/Δefg1 cell survival is unrelated to ROS production.
FIG 4.
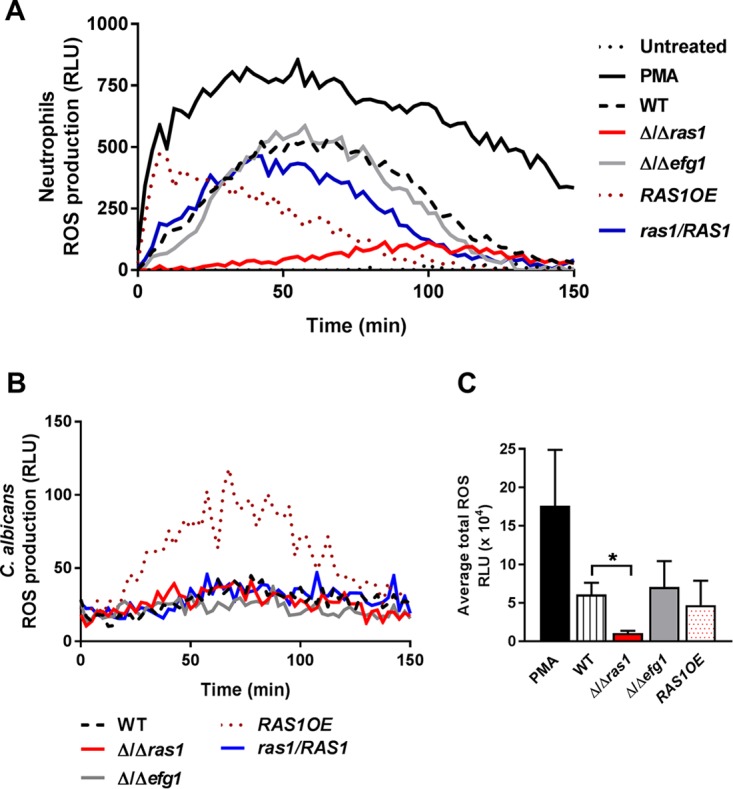
C. albicans Δ/Δras1 suppressed ROS production. (A) Human neutrophil ROS production upon coincubation with C. albicans WT, Δ/Δras1, Δ/Δefg1, RAS1OE, and ras1/RAS1 complemented strains was detected with luminol chemiluminescence over 150 min at 37°C. As controls, neutrophils were left untreated (negative) or treated with PMA (positive). The graph is a representation of one biological replicate. (B) C. albicans WT, Δ/Δras1, Δ/Δefg1, RAS1OE, and ras1/RAS1 complemented strain ROS production was detected using luminol as indicated for panel A. (C) Total ROS production was obtained by calculating the area under the curve of four independent experiments performed in duplicate. Shown are the means ± SDs from four independent experiments performed in duplicate. Significance was calculated using one-way ANOVA with post hoc Dunnett’s multiple-comparison test. *, P < 0.05.
C. albicans Δ/Δras1 cells have a higher tolerance to oxidative stress through increased catalase activity.
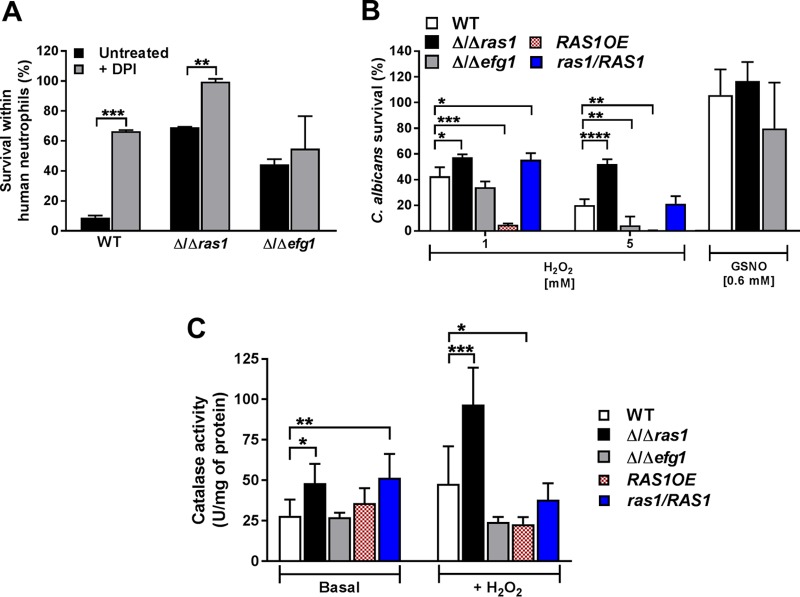
To demonstrate the importance of ROS production in C. albicans killing within neutrophils, we treated H-PMNs with the NADPH oxidase inhibitor diphenyleneidonium chloride (DPI) before adding C. albicans, expecting to see an increase in survival of C. albicans strains when ROS production was blocked (see Fig. S1 in the supplemental material). Indeed, we observed that upon ROS inhibition, C. albicans WT and Δ/Δras1 strain survival rates were significantly increased (66.4% and 99.6%, respectively) compared to those in untreated neutrophils (10% and 69.2%). However, there was still a difference in survival between WT and Δ/Δras1 cells within DPI-treated neutrophils (Fig. 5A), suggesting that changes in survival between the strains were only partially due to increased oxidative stress resistance. Furthermore, an increase in survival of the Δ/Δefg1 mutant within DPI-treated neutrophils was not observed, further suggesting that the Δ/Δefg1 mutant survival is governed by mechanisms other than ROS resistance (Fig. 5A). Therefore, we compared the survival of C. albicans Δ/Δras1 and Δ/Δefg1 cells upon exposure to both oxidative and nitrosative stresses. Since low levels of intracellular cAMP in C. albicans have been associated with increased expression of genes for resistance to oxidative stress, including catalase genes (38), we expected that Δ/Δras1, but not Δ/Δefg1, cells would be more resistant to hydrogen peroxide (H2O2). Indeed, C. albicans Δ/Δras1 cells were significantly more resistant to H2O2 at both 1 mM and 5 mM than the WT, while C. albicans Δ/Δefg1 cells had a susceptibility similar to that of the WT at 1 mM but higher susceptibility at 5 mM (Fig. 5B). Additionally, the effect observed in the absence of Ras1 was restored in the complemented ras1/RAS1 strain, which showed a survival rate similar to that of the WT at 5 mM (survival rates of 21% and 20%, respectively). Moreover, C. albicans RAS1OE cells were significantly less resistant to H2O2 at both concentrations tested (Fig. 5B). Cell sensitivity to nitrosative stress using S-nitrosoglutathione (GSNO) was also tested; however, all strains were completely resistant to this nitrosative stress after 2 h of treatment (Fig. 5B). Thus, ROS, but not RNS, are important determinants for survival of C. albicans Δ/Δras1 within neutrophils.
FIG 5.
C. albicans Δ/Δras1 increased resistance to oxidative stress. (A) Human blood-derived neutrophils were treated with diphenyleneiodonium chloride (DPI) for 15 min at 37°C, followed by infection with C. albicans WT, Δ/Δras1, or Δ/Δefg1 strains for 3 h. H-PMNs were lysed and internalized C. albicans cells were released, collected, and plated on agar for 48 h to obtain CFU. Survival was calculated as follows: (recovered C. albicans CFU after H-PMN lysis/total number of phagocytosed C. albicans cells) × 100. Shown are the means ± SDs from three independent experiments performed in duplicate. (B) C. albicans strains were treated with the indicated concentrations of H2O2 or S-nitrosoglutathione (GSNO) for 2 h at 30°C. Control cells were left untreated. After incubation, C. albicans cells were diluted and plated on agar, and CFU were obtained after 48 h. Survival was calculated as follows: (CFU of treated plate/CFU control plate) × 100. Shown are the means ± SDs from three independent experiments performed in duplicate. (C) Catalase activity was measured colorimetrically at 570 nm using whole-cell extracts isolated from mid-log-phase cells incubated for 1 h at 30°C without (basal) or with 5 mM H2O2. Shown are means ± SDs from three independent experiments performed in duplicate. Significance was calculated using unpaired Student t tests (A) or one-way ANOVA with post hoc Dunnett’s multiple-comparison tests (B and C). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
C. albicans is able to induce catalase expression as a mechanism to detoxify ROS and resist phagocytic killing (39, 40), and catalase levels are higher in cells with reduced levels of cAMP (31, 41). Therefore, we hypothesized that C. albicans Δ/Δras1 cells would have higher basal catalase expression and an increased catalase response to H2O2-mediated oxidative stress compared to the WT. As expected, C. albicans Δ/Δras1 had 2-fold-higher levels of basal catalase activity as well as a 2-fold increase in activity in response to H2O2 compared to the WT (Fig. 5C). In contrast, C. albicans Δ/Δefg1 mutant levels of basal and induced catalase activity were similar to those of WT cells, showing that higher catalase activity is specific to C. albicans Δ/Δras1 cells. Furthermore, C. albicans overexpressing Ras1 had decreased catalase activity under treatment compared to that of the WT (Fig. 5C).
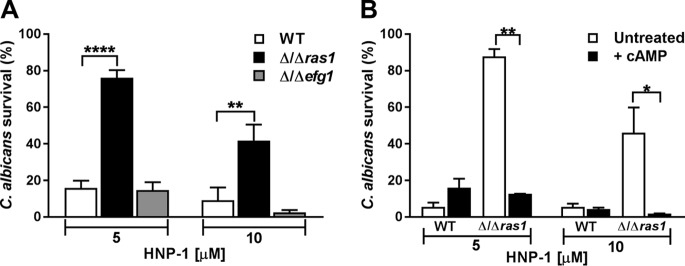
C. albicans Δ/Δras1 cell resistance to HNP-1 was reversed by cAMP.
In addition to oxidative mechanisms, human neutrophils utilize nonoxidative antimicrobial peptides localized within primary granules to kill pathogenic microorganisms. HNP-1 is one of the most abundant cationic peptides within granules of human neutrophils, but it is not present in murine neutrophils (42). To examine nonoxidative killing of C. albicans Δ/Δras1 cells by HNP-1, we first compared the in vitro susceptibility of C. albicans WT and Δ/Δefg1 cells. Surprisingly, 76.1% of C. albicans Δ/Δras1 cells survived HNP-1 treatment (5 μM), compared to only 15.8% of WT cells, and treatment of yeast with 10 μM HNP-1 resulted in 9.2% survival of WT cells, compared to 41.7% of Δ/Δras1 cells (Fig. 6A). C. albicans Δ/Δefg1 cells showed survival rates similar to those of the WT following treatment at both HNP-1 concentrations.
FIG 6.
C. albicans Δ/Δras1 cells have increased resistance to human neutrophil peptide 1 (HNP-1) via a cAMP-dependent mechanism. (A) C. albicans WT, Δ/Δras1, and Δ/Δefg1 strains were treated with 5 or 10 μM human neutrophil peptide 1 for 1 h at 37°C. Cells were diluted in NaPB, plated on agar, and incubated for 48 h to obtain viable CFU. (B) C. albicans WT and Δ/Δras1 strains were pretreated with 10 mM cAMP during exponential growth. After pretreatment, cells were incubated with HNP-1 as described above. Control cells were left untreated for all experiments. Survival was calculated as follows: (CFU of treated plate/CFU control plate) × 100. Shown are means ± SDs from three independent experiments performed in duplicate. Significance was calculated using one-way ANOVA with post hoc Dunnett’s multiple-comparison tests (A) or unpaired Student t test (B). *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Since low levels of ATP have been associated with an increased rigidity of the Candida cell membrane, making it less susceptible to the action of cationic antimicrobial peptides (32), we hypothesized that increasing cAMP levels in C. albicans Δ/Δras1 cells would restore killing by HNP-1. As predicted, pretreatment of C. albicans Δ/Δras1 cells with cAMP completely restored HNP-1 killing to levels of WT cells at both tested HNP-1 concentrations (Fig. 6B).
DISCUSSION
C. albicans MAP kinases and Ras1 signaling pathways are important contributors to virulence in mouse models of candidiasis (16, 24, 25) and are required for adaptation and resistance to a variety of stress conditions. To identify which of these MAP kinases contribute to C. albicans survival in phagocytes, we systematically tested for defects in phagocytosis and survival of C. albicans knockouts of individual kinases. We found that human neutrophils had significantly reduced phagocytic indices for C. albicans Δ/Δhog1 and Δ/Δras1 cells, suggesting that these mutants have significant alterations in surface cell wall composition involved in innate immune cell recognition. Although Δ/Δcek1 cells have alterations in cell wall composition (26), we did not find a measurable reduction in phagocytosis, contrary to previous observations by Galán-Díez et al. (26).
We next tested for survival of C. albicans cells that were taken up by neutrophils, independent from their rate of phagocytosis. As expected from their established role in protection from oxidative stress (43) and cell wall stress defense (44), HOG1 and CEK1 knockouts had reduced survival in neutrophils. C. albicans Δ/Δmkc1 cells, in contrast, had phagocytosis and survival similar to those of WT cells, thus showing that the primary role of MKC1 is unrelated to recognition and survival within immune cells. Unexpectedly, C. albicans Δ/Δras1 cell survival within the phagosome was increased to 106% within human primary neutrophils, suggesting that Δ/Δras1 cells not only survived but also potentially replicated within human neutrophils. To our knowledge, this is the only example of a C. albicans gene knockout resulting in complete survival of yeast within neutrophils.
Ras1 and Cyr1 are major regulators of fungal cellular cAMP levels, and these, in turn, control the expression of genes required for stress defense. Low levels of cAMP are linked with increased fungal survival following environmental oxidative stress, while high levels of cAMP have the opposite effect (30, 31, 38). Since C. albicans Δ/Δras1 cells have decreased intracellular cAMP (41), it is likely that altered cAMP levels in Δ/Δras1 contribute to increased survival within neutrophils. This seems not to be the case for C. albicans Δ/Δcyr1 cells, which showed no improvement in survival compared to the WT despite very low cellular cAMP levels; however, these cells additionally have decreased resistance to osmotic/oxidative stresses, which may counteract any increased resistance to cell wall stressors and apoptosis (22, 31, 41, 45).
We also found that knockout of EFG1 resulted in significantly improved survival of C. albicans in human neutrophils. It is not known whether C. albicans EFG1 impacts cAMP levels or Ras1 activity, though we did not observe increased resistance to oxidative or nonoxidative mechanisms that could explain this phenotype. The resistance to phagosomal killing of C. albicans Δ/Δefg1 cells could be based on the high redundancy in pathways regulated by Efg1 (46) and its complex regulatory network; however, this remains to be elucidated. In any case, its resistance to neutrophil killing appears to be through mechanisms other than those provided by Ras1.
Oxidative molecules released within the phagosome are crucial for killing engulfed pathogens, and we found that when ROS production was blocked by an NADPH oxidase inhibitor, survival of WT yeast cells increased 7-fold, but C. albicans Δ/Δras1 cells survival increased only 30% (Fig. 5A), suggesting that ROS contributed only about a third of the array of neutrophil killing mechanisms for these cells, compared with up to 80% for WT cells. Other crucial mechanisms for C. albicans Δ/Δras1 cell survival are suppression of ROS release by human neutrophils (Fig. 4A), increased catalase activity (Fig. 5C), and significant resistance to HNP-1 (Fig. 6A). In addition to inhibiting ROS release by neutrophils, as low cAMP levels stimulate fungal superoxide dismutase (SOD) production (38), it is possible that the lower cAMP levels in C. albicans Δ/Δras1 cells might increase the rate of ROS detoxification, rather than actual generation of phagosomal ROS. Alternatively, defective phagocytosis of C. albicans Δ/Δras1 cells (Fig. 2A) might affect the assembly of NOX2 and subsequent phagosome maturation within neutrophils, as C. albicans is capable of altering phagosome maturation and reducing ROS release in macrophages in order to resist killing (12). Therefore, whether the increased catalase production by C. albicans Δ/Δras1 we saw is sufficient to cause the reduction in detectable ROS production from neutrophils remains to be determined.
Most remarkable is our finding that C. albicans Δ/Δras1 cells displayed significant resistance to HNP-1 killing, since this antimicrobial peptide is generally considered to be a nonspecific membrane lytic peptide with a broad spectrum of activity against both bacteria and fungi. Although there is an association between ATP and cAMP levels of target microorganisms with their resistance to other antimicrobial peptides, such as histatin 5 (32, 47), this is this first evidence that C. albicans Ras1 can blunt the effects of HNP-1. We suggest a mechanism of fungal killing by HNP-1 that is dependent upon cellular cAMP, since inactivation of Ras1 which is linked to ATP and cAMP levels (19) reduced killing, while killing was completely restored upon replacement of cAMP. Furthermore, low ATP levels in C. albicans alter both the cell membrane state and entry of antifungal peptides (32), suggesting that the fungicidal mechanism of HNP-1 is dependent upon the energy status of the cell and requires active transport of HNP-1 rather than being a non-energy-requiring membrane-lytic peptide.
Downregulation of C. albicans Ras1 expression is an important mechanism that may be used by fungal cells to survive within phagocytes. Other pathogenic organisms resist intraphagosomal killing and then use immune cells as a vehicle to disseminate infection (48, 49). Indeed, the yeast Cryptococcus neoformans resists macrophage killing (50) and then uses phagocytes to disseminate to other body sites (51), as illustrated by decreased dissemination during infection upon phagocytic depletion (52). It is possible that C. albicans employs similar mechanisms in which Ras1 downregulation reduces its phagosome killing and allows fungal dissemination through neutrophils. Thus, Ras1 may be a key modulator of fungal cell survival within the host, and this pathway needs further study to understand its role in C. albicans infection.
MATERIALS AND METHODS
C. albicans strains and culture conditions.
C. albicans strains genotypes are indicated in Table 1. The C. albicans CAI-4 wild type (WT) and Δ/Δcek1 mutant were kindly provided by Malcolm Whiteway (Concordia University, Montreal, Canada), the Δ/Δhog1 mutant was provided by Janet Quinn (Newcastle University, Newcastle, United Kingdom), the Δ/Δmkc1 mutant was provided by Carol Kumamoto (Tuft University), the Δ/Δras1 and ras1/ras1-G13V (RAS1OE) mutants and ras1/RAS1 complemented strain were provided by Deborah Hogan (Dartmouth College), and the Δ/Δcyr1 and Δ/Δefg1 mutants were provided by Amy Piispanen (Dartmouth College). All C. albicans strains were cultured for 12 h in yeast extract-peptone-dextrose medium (YPD; BD Difco) broth supplemented with 50 μg/ml of uridine (Sigma-Aldrich) at 30°C in an orbital shaker at 220 rpm. Cultures were diluted to an optical density at 600 nm (OD600) of 0.3 to 0.4 in fresh YPD medium and then recultured to an OD600 of 0.7 to 0.8. C. albicans cells were pelleted by centrifugation at 2,500 × g for 5 min, washed thrice with phosphate-buffered saline (PBS; pH 7.4; Corning), and suspended in RPMI 1640 (Corning) or other indicated media. To induce hypha formation, C. albicans yeast (OD600 = 0.3 to 0.4) were cultured in fresh yeast nitrogen base (YNB; MP Biomedicals) supplemented with 1.25% N-acetylglucosamine (GlcNAc; Sigma-Aldrich) and incubated for 2 to 4 h at 37°C. Cell suspensions were vortexed vigorously to avoid clumping, which was confirmed microscopically. Hyphal length was assessed in a Zeiss Axio Observer Z1 inverted fluorescence microscope (Carl Zeiss, Germany) and measured using ZEN 2011 (blue edition) software.
TABLE 1.
C. albicans strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| CAI-4 (wild type) | ura3Δ::imm434/URA3 | 56 |
| Δ/Δcek1 | ura3Δ::imm434/ura3Δ::imm434 cek1Δ::hisG-URA-hisG/cek1Δ::hisG | 16 |
| Δ/Δhog1 | Δura3::imm434/Δura3::imm434 his1::hisG/his1::hisG hog1::loxP-ura3-loxP/hog1::loxP-HIS1-loxP CIp20 (URA3 HIS1) | 57 |
| Δ/Δmkc1 | CAI-4, mkc1D::hisG/mkc1 D::hisG mkc1::pCK70 (URA3) | 58 |
| Δ/Δras1 | ura3::λimm434/ura3::λimm434 ras1::hisG/ras1::hisG::URA3 | 23 |
| Δ/Δcyr1 | ura3D::kimm434/ura3D::k imm434 cyr1D::hisG::cyr1 D::hisG | 59 |
| Δ/Δefg1 | ura3::kimm434/ura3::kimm434efg1::hisG/efg1::hisAhisGura3::kimm434/ura3::kimm434efg1::hiG | 60 |
| ras1/RAS1 | ura3::λimm434/ura3::λimm434 ras1::hisG/ras1::hisG::RAS1-URA3 | 61 |
| ras1/ras1-G13V (RAS1OE) | ura3Δ::λimm434/ura3Δ::λimm434 ras1::hisG/ras1::hisG::ras1-G13V-URA3 | 61 |
Isolation of human neutrophils.
Human blood-derived neutrophils (H-PMNs) were isolated from venous blood, collected into VACUETTE EDTA tubes (Greiner Bio-One) from healthy donors that had provided informed consent as approved by the University at Buffalo institutional review board (IRB; protocol 626714). Cells were isolated by density gradient centrifugation using 1-step Polymorphs (Accurate Chemicals & Scientific Corporation) following the manufacturer’s instructions. H-PMNs were suspended at a concentration of 2 × 106/ml in RPMI 1640 supplemented with l-glutamine (Corning) and 10% fetal bovine serum (FBS; Seradigm) and used immediately. Purity was confirmed by Wright-Giemsa (Polysciences, Inc.) and 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) staining.
Isolation of mouse neutrophils.
Isolation of mouse bone marrow-derived neutrophils (M-PMNs) was performed as previously described, with modifications (53). Long bones from 6- to 8-week-old female C57BL/6J mice (Jackson Laboratory) were collected and washed in α-MEM medium (Gibco, Life Technologies) to remove any tissue debris. Bone marrow was flushed out, washed by centrifugation at 800 × g for 5 min, and suspended in 1 ml of α-MEM medium. The cell suspension was carefully layered on top of a discontinuous Percoll gradient (55%, 60%, and 80%) and centrifuged at 1,200 × g for 30 min at 4°C. The lower band (M-PMN) was collected and cells were washed with Hanks’ balanced salt solution (HBSS) without calcium (Corning) at 800 × g for 5 min. Red blood cell lysis was achieved using red blood cell lysis buffer (BioLegend) for 5 min at room temperature (RT; 21°C) with gentle shaking. After a final wash with HBSS without calcium, cells were resuspended in RPMI 1640 at a density of 2 × 106/ml. Cell viability was evaluated by trypan blue (Sigma-Aldrich) exclusion, and purity was determined by Wright-Giemsa and DAPI staining.
Evaluation of C. albicans uptake by PI.
For phagocytosis assays, H-PMNs and M-PMNs were seeded in 12-well plates and C. albicans resuspended in RMPI 1640 to an OD600 of 0.7 to 0.8 was added at a multiplicity of infection (MOI) of 3 and incubated for 30 min at 37°C (in a 5% CO2 environment) to allow phagocytosis. Nonadherent neutrophils were collected after C. albicans phagocytosis and placed on positively charged slides (Globe Scientific Inc.) for 15 min at RT to allow cells to attach. Following attachment, cells were gently washed with ice-cold PBS to remove medium, and 4 mg/ml of calcofluor white in PBS (CW; Sigma-Aldrich) was added for 2 min on ice to stain nonphagocytosed C. albicans. Next, cells were washed with ice-cold PBS to remove excess CW and fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 30 min at RT. After fixation, cells were permeabilized with 0.1% Triton X-100 (Fisher Bioreagents) for 5 min and stained with 4 mg/ml of Alexa Fluor 488-conjugated phalloidin (Invitrogen) for 5 min. After a final wash, positively charged slides were covered with number 1 cover glass (Knittel Glaser) using fluorescent mounting medium (Dako). Cells were counted using a Zeiss Axio Observer Z1 inverted fluorescence microscope (Carl Zeiss, Germany). A minimum of 100 phagocytic cells were observed for each experiment, and C. albicans cells that were partially or not stained with CW were counted as phagocytosed. The phagocytic index (PI) was calculated as the ratio of the total number of phagocytosed C. albicans cells and the total number of neutrophils counted. Assays were performed in duplicates, and experiments were repeated at least thrice.
Intracellular survival assay of C. albicans strains within neutrophils.
Survival assays were performed following the protocol described previously (54). Briefly, fungal and phagocytic cells were incubated in RPMI 1640 medium for 3 h at 37°C and 5% CO2 at an MOI of 0.1. This MOI was used to ensure that approximately 100% of added C. albicans WT cells were phagocytosed, as determined by microscopy PI determination as described above. The total number of phagocytosed C. albicans cells was calculated by multiplying the PI by the total number of neutrophils added. After 3 h of incubation, sterile water and 0.25% SDS (Thermo Fisher Scientific) were added to lyse neutrophils and release phagocytosed C. albicans. Cell suspensions were collected, vortexed vigorously to avoid cell clumping, and diluted. Finally, C. albicans cells were plated on YPD BD Difco agar and incubated for 48 h at 30°C to obtain viable CFU. Percent survival was determined as follows: (recovered C. albicans CFU after phagocytic cell lysis/total number of phagocytosed C. albicans) × 100. To block ROS release, phagocytic cells were treated before addition of C. albicans with 20 μM diphenyleneiodonium chloride (DPI; Sigma-Aldrich) for 15 min at 37°C and 5% CO2 (55). Assays were performed in duplicates, and results are representative of those from at least three independent experiments.
ROS production assay.
ROS production by H-PMNs was evaluated as described previously, with minor modifications (36). Human neutrophils (0.5 × 106/ml) were resuspended in RPMI 1640 without phenol red, supplemented with 5% FBS, and seeded (100 μl) in a 96-well plate (Corning Inc.). To avoid nonspecific activation, the plate was treated with 0.05% albumin (Sigma-Aldrich) at 4°C for 1 h and then washed twice with 200 μl of Dulbecco’s phosphate-buffered saline (DPBS; Corning Inc.) prior to seeding of neutrophils. C. albicans strains were grown as described above, resuspended in RPMI 1640 at a concentration of 5 × 106/ml, and then added to each well at an MOI of 5. A control for each strain was performed without neutrophils. Immediately after addition of C. albicans, luminol (50 μl of 200 μM stock) and 16 U of horseradish peroxidase (HRP) (both from Sigma-Aldrich) were added to wells to detect total ROS production. Luminescence was measured using an integration time of 1 s at intervals of 2.5 min over 2.5 h at 37°C using a FlexStation 3 multimode microplate reader (Molecular Devices). Relative light units (RLU) were obtained by subtracting the luminescence of the wells that contained only C. albicans without neutrophils from luminescence in experimental wells. Unstimulated and PMA-stimulated (750 ng/ml) neutrophils were used as negative and positive controls, respectively.
Oxidative and nonoxidative resistance assays.
For cAMP experiments, overnight cultures of C. albicans were diluted to an OD600 of 0.3 to 0.4 in YPD medium was supplemented with 10 mM dibutyryl-cAMP (Sigma-Aldrich) and then cultured to an OD600 of 0.7 to 0.8. C. albicans cells were then washed three times with 10 mM sodium phosphate buffer, pH 7.4 (NaPB), and then the cells (1.5 × 106/ml) were treated with 5 μM or 10 μM HNP-1 (AnaSpec Inc.) for 1 h at 37°C. After treatment, cells were serially diluted in NaPB and plated on YPD, and surviving cells were counted after incubation for 48 h at 30°C. Oxidative stress resistance was tested by incubation of C. albicans cells (1 × 107/ml) in YPD medium supplemented with 1 mM or 5 mM H2O2 (Sigma-Aldrich) or 0.6 mM S-nitrosoglutathione (GSNO; Sigma) for 2 h at 30°C with shaking. Percentage of survival was calculated as follows: (CFU from treated plate/CFU from untreated control plate) × 100.
Catalase assay.
Catalase activity was measured using an EnzyChrom catalase assay kit (BioAssay Systems). C. albicans WT and mutant strains cultured as described above were incubated with or without 5 mM H2O2 for 1 h at 30°C and then harvested, and catalase activity was measured colorimetrically according to the manufacturer’s instructions.
Statistical analysis.
All data were analyzed by GraphPad Prism software version 7 (GraphPad Software, San Diego, CA), and a significance level (α) of 0.05 was used for all experiments.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by NIH-NIDCR awards R01DE010641 and R01DE022720 (to M.E.), R03DE025062 (to J.G.K.), and CONICYT- Chile Scholarship 72150173 (to O.S.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00685-18.
REFERENCES
- 1.Norris HL, Friedman J, Chen Z, Puri S, Wilding G, Edgerton M. 2018. Salivary metals, age, and gender correlate with cultivable oral Candida carriage levels. J Oral Microbiol 10:1447216. doi: 10.1080/20002297.2018.1447216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trautwein-Weidner K, Gladiator A, Nur S, Diethelm P, LeibundGut-Landmann S. 2015. IL-17-mediated antifungal defense in the oral mucosa is independent of neutrophils. Mucosal Immunol 8:221–231. doi: 10.1038/mi.2014.57. [DOI] [PubMed] [Google Scholar]
- 3.Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang C-H, Webster KM. 2009. Epidemiology and outcomes of Candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis 48:1695–1703. doi: 10.1086/599039. [DOI] [PubMed] [Google Scholar]
- 4.Qian Q, Jutila MA, Van Rooijen N, Cutler JE. 1994. Elimination of mouse splenic macrophages correlates with increased susceptibility to experimental disseminated candidiasis. J Immunol 152:5000–5008. [PubMed] [Google Scholar]
- 5.Rudkin FM, Bain JM, Walls C, Lewis LE, Gow NA, Erwig LP. 2013. Altered dynamics of Candida albicans phagocytosis by macrophages and PMNs when both phagocyte subsets are present. mBio 4:e00810-13. doi: 10.1128/mBio.00810-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vonk AG, Wieland CW, Netea MG, Kullberg BJ. 2002. Phagocytosis and intracellular killing of Candida albicans blastoconidia by neutrophils and macrophages: a comparison of different microbiological test systems. J Microbiol Methods 49:55–62. doi: 10.1016/S0167-7012(01)00348-7. [DOI] [PubMed] [Google Scholar]
- 7.Ermert D, Niemiec MJ, Rohm M, Glenthoj A, Borregaard N, Urban CF. 2013. Candida albicans escapes from mouse neutrophils. J Leukoc Biol 94:223–236. doi: 10.1189/jlb.0213063. [DOI] [PubMed] [Google Scholar]
- 8.Dupré-Crochet S, Erard M, Nüβe O. 2013. ROS production in phagocytes: why, when, and where? J Leukoc Biol 94:657–670. doi: 10.1189/jlb.1012544. [DOI] [PubMed] [Google Scholar]
- 9.Nordenfelt P, Tapper H. 2011. Phagosome dynamics during phagocytosis by neutrophils. J Leukoc Biol 90:271–284. doi: 10.1189/jlb.0810457. [DOI] [PubMed] [Google Scholar]
- 10.Pethe K, Swenson DL, Alonso S, Anderson J, Wang C, Russell DG. 2004. Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc Natl Acad Sci U S A 101:13642–13647. doi: 10.1073/pnas.0401657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flannagan RS, Cosio G, Grinstein S. 2009. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol 7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Arenas E, Bleck CK, Nombela C, Gil C, Griffiths G, Diez-Orejas R. 2009. Candida albicans actively modulates intracellular membrane trafficking in mouse macrophage phagosomes. Cell Microbiol 11:560–589. doi: 10.1111/j.1462-5822.2008.01274.x. [DOI] [PubMed] [Google Scholar]
- 13.Monge RA, Román E, Nombela C, Pla J. 2006. The MAP kinase signal transduction network in Candida albicans. Microbiology 152:905–912. doi: 10.1099/mic.0.28616-0. [DOI] [PubMed] [Google Scholar]
- 14.Roman E, Arana DM, Nombela C, Alonso-Monge R, Pla J. 2007. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol 15:181–190. doi: 10.1016/j.tim.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Ramírez-Zavala B, Weyler M, Gildor T, Schmauch C, Kornitzer D, Arkowitz R, Morschhäuser J. 2013. Activation of the Cph1-dependent MAP Kinase signaling pathway induces white-opaque switching in Candida albicans. PLoS Pathog 9:e1003696. doi: 10.1371/journal.ppat.1003696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Csank C, Schroppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas DY, Whiteway M. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun 66:2713–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernst JF, Pla J. 2011. Signaling the glycoshield: maintenance of the Candida albicans cell wall. Int J Med Microbiol 301:378–383. doi: 10.1016/j.ijmm.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Alonso-Monge R, Navarro-Garcia F, Roman E, Negredo AI, Eisman B, Nombela C, Pla J. 2003. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot Cell 2:351–361. doi: 10.1128/EC.2.2.351-361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grahl N, Demers EG, Lindsay AK, Harty CE, Willger SD, Piispanen AE, Hogan DA. 2015. Mitochondrial activity and Cyr1 are key regulators of Ras1 activation of C. albicans virulence pathways. PLoS Pathog 11:e1005133. doi: 10.1371/journal.ppat.1005133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollomon JM, Grahl N, Willger SD, Koeppen K, Hogan DA. 2016. Global role of cyclic AMP signaling in pH-dependent responses in Candida albicans. mSphere 1:e00283-16. doi: 10.1128/mSphere.00283-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inglis DO, Sherlock G. 2013. Ras signaling gets fine-tuned: regulation of multiple pathogenic traits of Candida albicans. Eukaryot Cell 12:1316–1325. doi: 10.1128/EC.00094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocha CRC, Schröppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell 12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leberer E, Harcus D, Dignard D, Johnson L, Ushinsky S, Thomas DY, Schroppel K. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol Microbiol 42:673–687. [DOI] [PubMed] [Google Scholar]
- 24.Diez-Orejas R, Molero G, Navarro-García F, Pla J, Nombela C, Sanchez-Pérez M. 1997. Reduced virulence of Candida albicans MKC1 mutants: a role for mitogen-activated protein kinase in pathogenesis. Infect Immun 65:833–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheetham J, MacCallum DM, Doris KS, da Silva Dantas A, Scorfield S, Odds F, Smith DA, Quinn J. 2011. MAPKKK-independent regulation of the Hog1 stress-activated protein kinase in Candida albicans. J Biol Chem 286:42002–42016. doi: 10.1074/jbc.M111.265231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galán-Díez M, Arana DM, Serrano-Gómez D, Kremer L, Casasnovas JM, Ortega M, Cuesta-Domínguez A, Corbí AL, Pla J, Fernández-Ruiz E. 2010. Candida albicans β-glucan exposure is controlled by the fungal CEK1-mediated mitogen-activated protein kinase pathway that modulates immune responses triggered through dectin-1. Infect Immun 78:1426–1436. doi: 10.1128/IAI.00989-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du C, Calderone R, Richert J, Li D. 2005. Deletion of the SSK1 response regulator gene in Candida albicans contributes to enhanced killing by human polymorphonuclear neutrophils. Infect Immun 73:865–871. doi: 10.1128/IAI.73.2.865-871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niemiec MJ, Grumaz C, Ermert D, Desel C, Shankar M, Lopes JP, Mills IG, Stevens P, Sohn K, Urban CF. 2017. Dual transcriptome of the immediate neutrophil and Candida albicans interplay. BMC Genomics 18:696. doi: 10.1186/s12864-017-4097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bahn YS, Staab J, Sundstrom P. 2003. Increased high‐affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1‐dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol Microbiol 50:391–409. doi: 10.1046/j.1365-2958.2003.03692.x. [DOI] [PubMed] [Google Scholar]
- 30.Wilson D, Tutulan-Cunita A, Jung W, Hauser NC, Hernandez R, Williamson T, Piekarska K, Rupp S, Young T, Stateva L. 2007. Deletion of the high-affinity cAMP phosphodiesterase encoded by PDE2 affects stress responses and virulence in Candida albicans. Mol Microbiol 65:841–856. doi: 10.1111/j.1365-2958.2007.05788.x. [DOI] [PubMed] [Google Scholar]
- 31.Harcus D, Nantel A, Marcil A, Rigby T, Whiteway M. 2004. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol Biol Cell 15:4490–4499. doi: 10.1091/mbc.e04-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veerman EC, Valentijn-Benz M, Nazmi K, Ruissen AL, Walgreen-Weterings E, van Marle J, Doust AB, van't Hof W, Bolscher JG, Amerongen AV. 2007. Energy depletion protects Candida albicans against antimicrobial peptides by rigidifying its cell membrane. J Biol Chem 282:18831–18841. doi: 10.1074/jbc.M610555200. [DOI] [PubMed] [Google Scholar]
- 33.Champion JA, Mitragotri S. 2006. Role of target geometry in phagocytosis. Proc Natl Acad Sci U S A 103:4930–4934. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal AW. 2005. How neutrophils kill microbes. Annu Rev Immunol 23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossi DCP, Gleason JE, Sanchez H, Schatzman SS, Culbertson EM, Johnson CJ, McNees CA, Coelho C, Nett JE, Andes DR, Cormack BP, Culotta VC. 2017. Candida albicans FRE8 encodes a member of the NADPH oxidase family that produces a burst of ROS during fungal morphogenesis. PLoS Pathog 13:e1006763. doi: 10.1371/journal.ppat.1006763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miramon P, Dunker C, Windecker H, Bohovych IM, Brown AJ, Kurzai O, Hube B. 2012. Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS One 7:e52850. doi: 10.1371/journal.pone.0052850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Zhao S, Sun L, Li W, Wei Q, Ashman RB, Hu Y. 2017. Different virulence of Candida albicans is attributed to the ability of escape from neutrophil extracellular traps by secretion of DNase. Am J Transl Res 9:50–62. [PMC free article] [PubMed] [Google Scholar]
- 38.Bahn YS, Molenda M, Staab JF, Lyman CA, Gordon LJ, Sundstrom P. 2007. Genome-wide transcriptional profiling of the cyclic AMP-dependent signaling pathway during morphogenic transitions of Candida albicans. Eukaryot Cell 6:2376–2390. doi: 10.1128/EC.00318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa Y, Koide K, Watanabe K, Morita Y, Mizuguchi I, Akashi T. 1999. The expression of the pathogenic yeast Candida albicans catalase gene in response to hydrogen peroxide. Microbiol Immunol 43:645–651. doi: 10.1111/j.1348-0421.1999.tb02452.x. [DOI] [PubMed] [Google Scholar]
- 40.Pradhan A, Herrero-de-Dios C, Belmonte R, Budge S, Lopez Garcia A, Kolmogorova A, Lee KK, Martin BD, Ribeiro A, Bebes A, Yuecel R, Gow NAR, Munro CA, MacCallum DM, Quinn J, Brown AJP. 2017. Elevated catalase expression in a fungal pathogen is a double-edged sword of iron. PLoS Pathog 13:e1006405. doi: 10.1371/journal.ppat.1006405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. 2008. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol 67:47–62. doi: 10.1111/j.1365-2958.2007.06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eisenhauer PB, Lehrer RI. 1992. Mouse neutrophils lack defensins. Infect Immun 60:3446–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arana DM, Alonso-Monge R, Du C, Calderone R, Pla J. 2007. Differential susceptibility of mitogen-activated protein kinase pathway mutants to oxidative-mediated killing by phagocytes in the fungal pathogen Candida albicans. Cell Microbiol 9:1647–1659. doi: 10.1111/j.1462-5822.2007.00898.x. [DOI] [PubMed] [Google Scholar]
- 44.Eisman B, Alonso-Monge R, Roman E, Arana D, Nombela C, Pla J. 2006. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell 5:347–358. doi: 10.1128/EC.5.2.347-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips AJ, Crowe JD, Ramsdale M. 2006. Ras pathway signaling accelerates programmed cell death in the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A 103:726–731. doi: 10.1073/pnas.0506405103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lane S, Birse C, Zhou S, Matson R, Liu H. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J Biol Chem 276:48988–48996. doi: 10.1074/jbc.M104484200. [DOI] [PubMed] [Google Scholar]
- 47.Jain P, Akula I, Edlind T. 2003. Cyclic AMP signaling pathway modulates susceptibility of Candida species and Saccharomyces cerevisiae to antifungal azoles and other sterol biosynthesis inhibitors. Antimicrob Agents Chemother 47:3195–3201. doi: 10.1128/AAC.47.10.3195-3201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gresham HD, Lowrance JH, Caver TE, Wilson BS, Cheung AL, Lindberg FP. 2000. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J Immunol 164:3713–3722. doi: 10.4049/jimmunol.164.7.3713. [DOI] [PubMed] [Google Scholar]
- 49.Kubica M, Guzik K, Koziel J, Zarebski M, Richter W, Gajkowska B, Golda A, Maciag-Gudowska A, Brix K, Shaw L, Foster T, Potempa J. 2008. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS One 3:e1409. doi: 10.1371/journal.pone.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levitz SM, Nong SH, Seetoo KF, Harrison TS, Speizer RA, Simons ER. 1999. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect Immun 67:885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.García-Rodas R, Zaragoza O. 2012. Catch me if you can: phagocytosis and killing avoidance by Cryptococcus neoformans. FEMS Immunol Med Microbiol 64:147–161. doi: 10.1111/j.1574-695X.2011.00871.x. [DOI] [PubMed] [Google Scholar]
- 52.Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F. 2009. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect Immun 77:120–127. doi: 10.1128/IAI.01065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ermert D, Urban CF, Laube B, Goosmann C, Zychlinsky A, Brinkmann V. 2009. Mouse neutrophil extracellular traps in microbial infections. J Innate Immun 1:181–193. doi: 10.1159/000205281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pathirana RU, Friedman J, Norris HL, Salvatori O, McCall AD, Kay J, Edgerton M. 2018. Fluconazole-resistant Candida auris is susceptible to salivary histatin 5 killing and to intrinsic host defenses. Antimicrob Agents Chemother 62:e01872-17. doi: 10.1128/aac.01872-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellis JA, Mayer SJ, Jones OT. 1988. The effect of the NADPH oxidase inhibitor diphenyleneiodonium on aerobic and anaerobic microbicidal activities of human neutrophils. Biochem J 251:887–891. doi: 10.1042/bj2510887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith DA, Nicholls S, Morgan BA, Brown AJP, Quinn J. 2004. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell 15:4179–4190. doi: 10.1091/mbc.e04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumamoto CA. 2005. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc Natl Acad Sci U S A 102:5576–5581. doi: 10.1073/pnas.0407097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindsay AK, Deveau A, Piispanen AE, Hogan DA. 2012. Farnesol and cyclic AMP signaling effects on the hypha-to-yeast transition in Candida albicans. Eukaryot Cell 11:1219–1225. doi: 10.1128/EC.00144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. doi: 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
- 61.Piispanen AE, Bonnefoi O, Carden S, Deveau A, Bassilana M, Hogan DA. 2011. Roles of Ras1 membrane localization during Candida albicans hyphal growth and farnesol response. Eukaryot Cell 10:1473–1484. doi: 10.1128/EC.05153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.