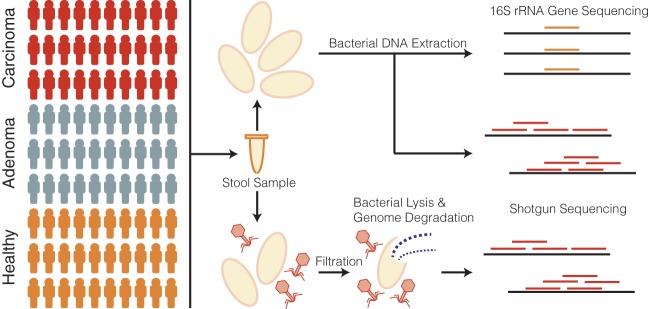
FIG 1.
Cohort and sample processing outline. Thirty subject stool samples were collected from healthy subjects and adenoma (precancer) and carcinoma (cancer) patients. Stool samples were split into two aliquots, the first of which was used for bacterial sequencing and the second of which was used for virus sequencing. Bacterial sequencing was done using both 16S rRNA amplicon and whole-metagenomic shotgun sequencing techniques. Virus samples were purified for viruses using filtration and a combination of chloroform (bacterial lysis) and DNase (exposed genomic DNA degradation). The resulting encapsulated virus DNA was sequenced using whole-metagenomic shotgun sequencing.