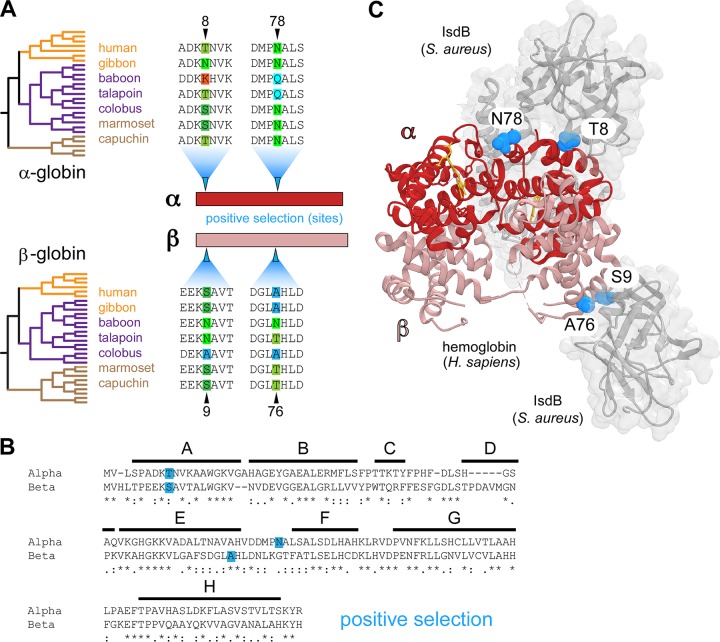
FIG 1.
Parallel signatures of positive selection among primate hemoglobins at the bacterial IsdB binding interface. (A) Species phylogenies for the 27 α-globin and 30 β-globin orthologs analyzed (left). Alignments from representative species across hominoid (orange), Old World monkey (purple), and New World monkeys (brown) are shown, the full phylogenetic trees are displayed in Fig. 3 and 4. Amino acid sites under positive selection were identified in α-globin and β-globin (blue arrows) using PAML with both species and gene trees (posterior probabilities, >0.95). (B) Amino acid alignment of human α-globin and β-globin proteins. The position of conserved helices A to H are shown, and identity between globins is noted as conserved (*), highly similar (:), and similar (.). (C) The residues of α-globin (red) and β-globin (salmon) under positive selection (blue spheres) at the interface of hemoglobin capture by Staphylococcus aureus IsdB (gray) (PDB 5VMM).