 Astaxanthin has been demonstrated to exhibit a wide range of beneficial effects that include anti-cancer and anti-inflammatory properties.
Astaxanthin has been demonstrated to exhibit a wide range of beneficial effects that include anti-cancer and anti-inflammatory properties.
Abstract
Astaxanthin has been demonstrated to exhibit a wide range of beneficial effects that include anti-cancer and anti-inflammatory properties. Xeroderma pigmentosum complementation group C (XPC) protein is an important DNA damage recognition factor in nucleotide excision repair and is involved in regulating non-small cell lung cancer (NSCLC) cell proliferation and viability. Erlotinib (TarcevaR) is a selective epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor that has demonstrated clinical activity in NSCLC cells. However, whether astaxanthin and erlotinib could induce synergistic cytotoxicity in NSCLC cells through modulating XPC expression is unknown. In this study, we found that p38 MAPK activation by astaxanthin decreased XPC expression in two human lung adenocarcinoma A549 and H1975 cells. Inactivation of p38 MAPK by pharmacological inhibitor SB203580 or the specific small interfering RNA (siRNA) rescued the astaxanthin-reduced XPC mRNA and protein levels. Enforced expression of XPC cDNA or inhibiting the p38 MAPK activity reduced the cytotoxicity and cell growth inhibition of astaxanthin. In contrast, knockdown of XPC using siRNA enhanced the cytotoxic effects of astaxanthin. Moreover, astaxanthin synergistically enhanced cytotoxicity and cell growth inhibition of erlotinib in NSCLC cells, which were associated with the down-regulation of XPC expression and activation of p38 MAPK. Our findings suggested that the astaxanthin induced p38 MAPK mediated XPC down-regulation enhanced the erlotinib-induced cytotoxicity in A549 and H1975 cells.
Introduction
Lung cancer is the most common cause of cancer-associated mortality in the world, and non-small cell lung cancer (NSCLC) accounts for the most lung cancer-related deaths.1–3 In NSCLC, the epidermal growth factor receptor (EGFR) is often overexpressed4 and EGFR signaling activation can enhance cell proliferation, anti-apoptosis, angiogenesis, and metastasis, leading to poor disease prognosis.5,6 Erlotinib, an EGFR tyrosine kinase inhibitor (TKI), has been approved to prolong survival in patients with advanced NSCLC after first-line and second-line chemotherapy.7 Previous studies have indicated that good objective tumor responses to EGFR-TKIs occur more frequently in patients who were women and have never smoked, and are higher in adenocarcinoma than other cancer types.8
In nucleotide excision repair pathways, xeroderma pigmentosum complementation group C (XPC) protein plays an important role in recognizing DNA damage.9 The XPC knockout mice are highly susceptible to UV radiation-induced skin cancer,10,11 and 2-acetylaminofluorene-induced lung and liver cancer.12 A previous study showed that genomic instability resulting from XPC silencing results in activation of AKT1 and NADPH oxidase-1 to induce reactive oxygen species generation and mitochondrial DNA deletions in human keratinocytes.13 Recently, Cui et al. revealed that XPC inhibits NSCLC cell proliferation and migration by enhancing E-Cadherin expression.14 XPC-deficient cells induce less apoptosis than their wild-type counterparts after UV irradiation, cisplatin, and etoposide.15 However, whether XPC expression is involved in erlotinib-induced cytotoxic effects in human lung cancer cells is unclear.
Astaxanthin, a ketocarotenoid (3,3′-dihydroxy-β,β′-carotene-4,4′-dione) red pigment, is widely distributed in nature, such as in microalgae, red yeast, shrimp, salmon, crayfish, and lobsters.16,17 Astaxanthin can effectively scavenge oxygen free radicals and reduce oxidative stress in both cell and animal models.18–20 Several health-promoting benefits have been associated with astaxanthin supplementation, including immunomodulation and prevention of cardiovascular diseases and cancer.19,21–24 Also, astaxanthin induces apoptosis in U937 cells (human leukemic monocyte lymphoma cell line) by downregulation of AKT activity.25 Recently, astaxanthin treatment was shown to promote apoptosis in dimethylhydrazine induced rat colon carcinogenesis by modulating the expressions of ERK1/2, NF-κB, and COX-2.26 However, whether astaxanthin could regulate XPC expression to affect erlotinib-induced cytotoxic effects in NSCLC has not been examined.
In this study, we wanted to explore the molecular mechanism of astaxanthin in regulating XPC expression to enhance the cytotoxic effect of erlotinib in human lung cancer cells. Using A549 and H1975 human lung adenocarcinoma cancer cell lines, we found that decreased XPC expression by astaxanthin was correlated with enhancing the sensitivity to erlotinib. These results may provide a rationale to combine astaxanthin with erlotinib for lung cancer treatment.
Materials and methods
Cell lines and chemicals
Human lung carcinoma cells A549 and H1975 were obtained from the American Type Culture Collection (Manassas, VA) and the cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2 in RPMI-1640 complete medium supplemented with sodium bicarbonate (2.2%, w/v), l-glutamine (0.03%, w/v), penicillin (100 units per mL), streptomycin (100 μg mL–1), and fetal calf serum (10%). The cell lines were routinely tested to confirm that they were free of Mycoplasma. Astaxanthin (purity ≥98%) was purchased from Sigma Chemical (St Louis, MO). Erlotinib was purchased from Genentech (South San Francisco, CA, USA). Cycloheximide and actinomycin D were purchased from Sigma-Aldrich (St Louis, MO, USA). MG132, lactacystin, and SB203580 were purchased from Calbiochem-Novabiochem (San Diego, CA, USA). Actinomycin D, lactacystin, MG132, and SB203580 were dissolved in dimethyl sulfoxide (DMSO). Cycloheximide was dissolved in Milli-Q-purified water (Millipore, Billerica, MA, USA).
Western blot analysis
After different treatments, equal amounts of proteins from each set of experiments were subjected to western blot analysis as previously described.27 The specific phospho-MKK3 (Ser189)/MKK6 (Ser207) and phospho-p38 MAPK (Thr180/Tyr182) antibodies were purchased from Cell Signaling (Beverly, MA, USA). Mouse monoclonal antibody against XPC(A-5) (sc-74411); rabbit polyclonal antibodies against p38(C-20) (sc-535), MKK3(N-20) (sc-959), actin(I-19) (sc-1616) and Ub(P4D1) (sc-8017) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The western blot shown was one representative of three independent experiments.
Plasmid and transfection
Plasmid transfections of MKK6E (a constitutively active form of MKK6) were achieved as previously described.28 Exponentially growing human lung cancer cells (106) were plated for 18 h, and then the MKK6E or Flag-XPC cDNA expression vectors were transfected into A549 or H1975 cells using Lipofectamine 2000 (Invitrogen). The sense-strand sequences of siRNA duplexes were as follows: the sense-strand sequences of small interfering RNA (siRNA) duplexes were as follows: p38: 5′-GAACUGCGGUUACUUAAAC-3′, 5′-AUGAAUGAUGGACUGAAAUGGUCUG-3′; and scrambled (as a control): 5′-UUCUCCGAACGUGUCACGUTT-3′ (Dharmacon Research, Lafayette, CO, USA). The XPC siRNA (sc-37805) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Cells were transfected with siRNA duplexes (100 nM) by using Lipofectamine 2000 (Invitrogen) for 24 h.
Quantitative real-time polymerase chain reaction (PCR)
PCRs were performed using an ABI Prism 7900HT according to the manufacturer's instructions. Amplification of specific PCR products was performed using the SYBR Green PCR Master Mix (Applied Biosystems). For each sample, the data were normalized to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The designed primers in this study were: XPC, forward, 5′-GACAAGCAG GAGAAGGCAAC-3′, reverse, 5′-GGTTCGGAATCCTCATCAGA-3′; GAPDH forward primer, 5′-CATGAGAAGTATGACAACAGCCT-3′; GAPDH reverse primer, 5′-AGTCCTTCCACGATACCAAAGT-3′. Analysis was performed using the comparative Ct value method. For each sample, the data were normalized to the housekeeping gene gapdh.
MTS assay
In vitro 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenol)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay was performed. Cells were cultured at 5000 per well in 96-well tissue culture plates. To assess cell viability, drugs were added after plating. At the end of the culture period, 20 μL of MTS solution (CellTiter 96 Aqueous One Solution Cell Proliferation Assay; Promega, Madison, WI, USA) was added, the cells were incubated for a further 2 h, and the absorbance was measured at 490 nm using an ELISA plate reader (Biorad Technologies, Hercules, CA).
Combination index analysis
Erlotinib and astaxanthin were combined at a ratio of 1 : 2, and the effect of combined treatment on cell viability was examined by MTS assay. To calculate a combination index (CI), the computer software Calcusyn (Biosoft, Oxford, UK) can be used, taking the entire shape of the cell viability curve into account for calculating whether a combination is synergistic (CI < 0.9), additive (CI = 0.9–1.1), or antagonistic (CI >1.1).29 The mean of CI values at a fraction affected (FA) of 0.90, 0.75, 0.50 were used to calculate between the three independent experiments.
Trypan blue dye exclusion assay
Cells were treated with astaxanthin and/or erlotinib for 24, 48, and 72 h. After treatment, the 500 cells were harvested, and the proportion of dead cells was determined by hemocytometer, performed by counting the number of cells stained with trypan blue. Trypan blue dye can be excluded from living cells but is able to penetrate dead cells. The dead cells were calculated as follows: trypan blue (+) cells ratio (%) = (stained cell number/total cell number) × 100.
Colony-forming ability assay
Immediately after drug treatment, the cells were washed with phosphate-buffered saline and trypsinized to determine the cell numbers. The cells were plated at a density of 500 cells on a 60 mm-diameter Petri dish in triplicate for each treatment and cultured for 12–14 days. The cell colonies were stained with 1% crystal violet solution in 30% ethanol. Cytotoxicity was determined by the number of colonies in the treated cells divided by the number of colonies in the untreated control.
Statistical analyses
For each protocol, three or four independent experiments were performed. Results were expressed as the mean ± SEM. Statistical calculations were performed using SigmaPlot 2000 software (Systat Software, San Jose, CA). Differences in measured variables between the experimental and control groups were assessed by unpaired t-test. P < 0.05 was considered statistically significant.
Results
XPC mRNA and protein levels were decreased after astaxanthin exposure
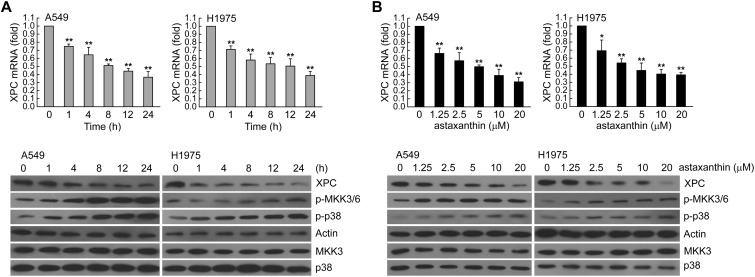
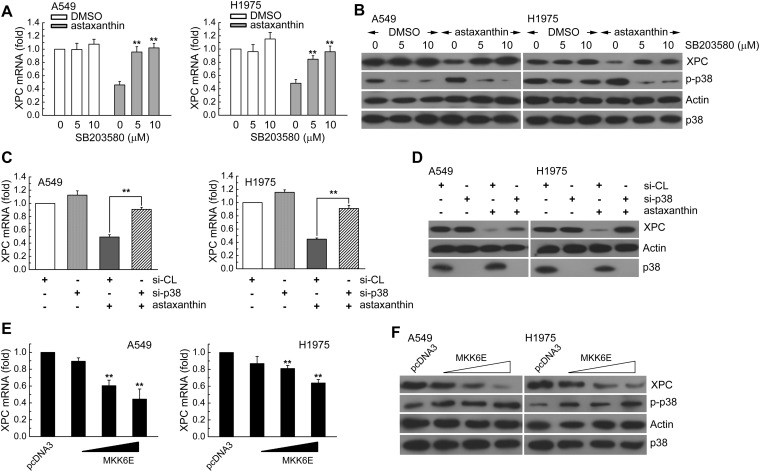
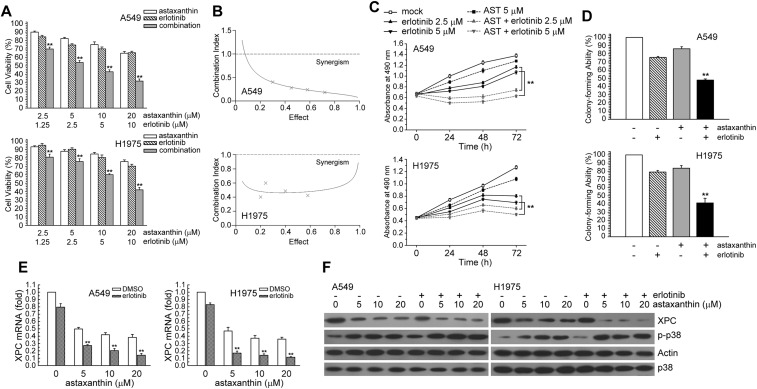
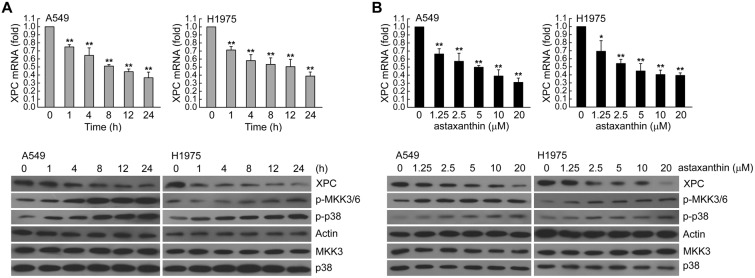
To determine whether XPC expression was associated with the effect of astaxanthin, we first assessed A549 or H1975 cells treated with astaxanthin (20 μM) for 1–24 h and examined various concentrations of astaxanthin (1.25, 2.5, 5, 10, 20 μM) for 24 h. Then, the real-time PCR and western blot analysis was used for determination of the XPC mRNA and protein level, respectively. In Fig. 1A and B, astaxanthin reduced XPC mRNA and protein expression in a time and dose-dependent manner; this was accompanied with an increase in phospho-MKK3/6 and phospho-p38 MAPK protein levels. To investigate the role of p38 MAPK activation in regulating XPC expression, we examined whether blockade of p38 MAPK activity could inhibit the down-regulation of XPC expression in astaxanthin-exposed A549 and H1975 cells. In Fig. 2A–D, these two cell lines were pretreated with p38 inhibitor (SB203580) or knockdown of p38 MAPK expression by specific siRNA transfection and then treated with astaxanthin. The results showed that inactivation of p38 MAPK by the pharmacological inhibitor or the specific siRNA rescued the astaxanthin-reduced XPC mRNA and protein levels (Fig. 2A–D). Furthermore, A549 or H1975 cells were transiently transfected with a plasmid carrying MKK6E, a constitutively active form of MKK6. Compared to transfection with the control vector-pcDNA3, transfection with MKK6E increased the p38 MAPK phosphorylation and decreased XPC mRNA and protein levels in A549 or H1975 cells (Fig. 2E and F). Therefore, astaxanthin down-regulated XPC expression occurred in a p38 MAPK activation manner.
Fig. 1. Astaxanthin decreased XPC expression in a dose and time-dependent manner. (A) Left panel, A549 or H1975 cells were exposed to astaxanthin (20 μM) for 1, 4, 8 12, or 24 h. Right panel, various concentrations of astaxanthin (1.25–20 μM) was added to cells for 24 h in complete medium. The total RNA was isolated and subjected to real-time PCR. The results (mean ± SEM) were from three independent experiments. *p < 0.05, **p < 0.01 using Student's t-test for comparison between the cells treated with or without astaxanthin. (B) After treatment as above, the cell extracts were examined by western blot analysis.
Fig. 2. Astaxanthin decreased XPC expression via MKK3/6-p38 MAPK activation in A549 and H1975 cells. (A and B) SB203580 (5 or 10 μM) was added to A549 or H1975 cells for 1 h before astaxanthin (5 μM) treatment for 24 h. The results (mean ± SEM) were from four independent experiments. **p < 0.01 using Student's t-test for comparison between the cells treated with astaxanthin–DMSO or a combination of astaxanthin–SB203580. (C and D) Cells were transfected with siRNA duplexes (200 nM) specific to p38 MAPK or scrambled (control) in complete medium for 24 h prior to treatment with astaxanthin (5 μM) in complete medium for 24 h. The results (mean ± SEM) were from 3 independent experiments. **p < 0.01, using Student's t-test for comparison between cells treated with astaxanthin in si-p38 MAPK RNA or si-scrambled RNA-transfected cells. (E and F) A549 or H1975 cells were transfected with MKK6E expression vector for 24 h. The results (mean ± SEM) were from 3 independent experiments. **p < 0.01 using Student's t-test for comparison of cells transfected with pcDNA3 and those transfected with MKK6E vector. After treatment, the cell extracts were examined via real-time PCR (A, C, E) and western blot (B, D, F) for determination of XPC mRNA and protein levels, respectively.
p38 MAPK activation by astaxanthin decreased XPC mRNA and protein stability
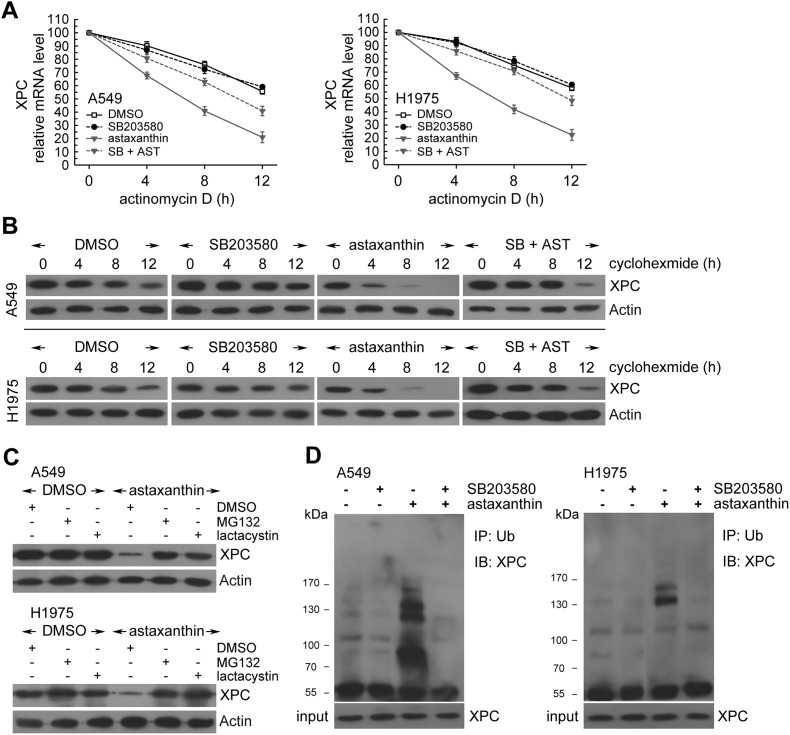
Next, we examined possible mechanisms for post-transcriptional regulation of XPC transcripts under astaxanthin treatment. In the presence of actinomycin D, astaxanthin treatment showed lower levels of XPC mRNA relative to untreated cells (Fig. 3A). Moreover, XPC protein levels were progressively reduced with time in the presence of cycloheximide (an inhibitor of de novo protein synthesis) (Fig. 3B). However, astaxanthin treatment significantly promoted XPC degradation after cycloheximide treatment compared to untreated cells (Fig. 3B). Therefore, XPC protein was less stable after astaxanthin treatment in A549 and H1975 cells. Interestingly, blocking p38 MAPK activation by SB203580 suppressed astaxanthin-induced XPC mRNA and protein instability (Fig. 3A and B). These results indicated that astaxanthin decreased XPC protein and mRNA levels through the improvement of protein and mRNA instability of XPC by p38 MAPK activation.
Fig. 3. Astaxanthin treatment decreased XPC mRNA and protein stability in a p38 MAPK activation manner. (A) Cells were treated with astaxanthin (20 μM) and/or SB203580 (10 μM) for 12 h in the presence or absence of actinomycin D (2 μg mL–1) for 4, 8, or 12 h; total RNA was isolated and subjected to real-time PCR. The results (mean ± SEM) were from 3 independent experiments. (B) The cells were exposed to astaxanthin (20 μM) and/or SB203580 (10 μM) for 12 h followed by co-treatment with cycloheximide (CHX; 0.1 mg mL–1) for 4, 8, or 12 h. Whole-cell extracts were collected for western blot analysis. (C) Astaxanthin (20 μM) was added to cells for 18 h, then the cells were co-treated with 26S proteasome inhibitor MG132 (10 μM) or lactacystin (10 μM) for 6 h. Whole cell extracts were collected for western blot analysis. (D) After pretreatment of SB203580 (10 μM) for 1 h, the cells were treated with astaxanthin (20 μM) for 18 h, and then MG132 (10 μM) for 6 h before whole cell extract collection. Equal amount of proteins in each cell extract were subjected to immunoprecipitation (IP) and western blotting analysis (IB).
Next, we hypothesized p38 MAPK activation to be involved in the down-regulation of XPC protein levels by the ubiquitin-26S proteasome-mediated proteolysis of XPC. In Fig. 3C, the 26S proteasome inhibitors MG132 or lactacystin were added for the final 6 h before harvesting in astaxanthin-treated A549 and H1975 cells. The results showed that both MG132 and lactacystin restored the decreased XPC protein levels induced by astaxanthin (Fig. 3C). In addition, we examined the levels of ubiquitin conjugates on XPC. Pretreatment of p38 MAPK inhibitor decreased the levels of ubiquitin-conjugated XPC in astaxanthin-treated NSCLC cell lines (Fig. 3D). Taken together, p38 MAPK activation decreased XPC protein levels by trigger XPC protein degradation via the ubiquitin-mediated 26S proteasome pathway in astaxanthin-treated NSCLC cells.
Down-regulation of XPC expression involved in regulating astaxanthin-induced cytotoxicity and growth inhibition in NSCLC cells
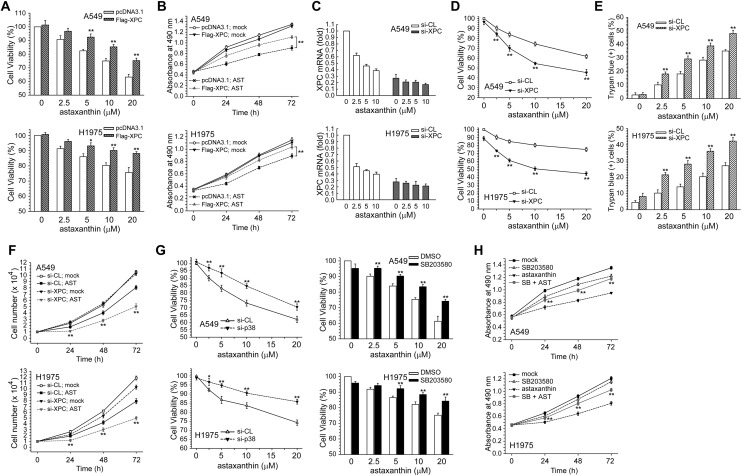
Next, the role of the decrease of XPC expression and p38 MAPK activation in the cytotoxic effect of astaxanthin was examined. In Fig. 4A and B, enforced expression of the Flag-XPC expression vector rescued A549 and H1975 cell viability after being decreased by astaxanthin, and also reversed the growth inhibition effect. We next examined the effect of siRNA-mediated XPC knockdown on astaxanthin-induced cytotoxicity and cell growth inhibition in NSCLC cells. At 24 h post-transfection, real-time PCR analysis showed a further decrease in XPC mRNA in astaxanthin-treated A549 and H1975 cells (Fig. 4C). Furthermore, suppression of XPC expression by si-XPC RNA resulted in increased sensitivity to astaxanthin compared to si-control transfected cells (Fig. 4D and E), and more inhibition of cell growth was induced by the combination of XPC siRNA and astaxanthin than by astaxanthin alone in A549 or H1975 cells (Fig. 4F). Therefore, down-regulation of XPC expression could enhance astaxanthin-induced cytotoxicity and growth inhibition in NSCLC cells.
Fig. 4. Enhancement of XPC expression by Flag-XPC cDNA vector transfection decreased the cytotoxicity induced by astaxanthin. (A) After the cells were transfected with pcDNA3 or Flag-XPC expression vector, the cells were treated with astaxanthin (2.5–20 μM μM) for 24 h. Cytotoxicity was determined by MTS assay. The results (mean ± SEM) were from three independent experiments. (B) After the cells were transfected with pcDNA3 or Flag-XPC expression vector, the cells were treated with astaxanthin (10 μM) for 24, 48, and 72 h, and cytotoxicity was determined by MTS assay. **p < 0.01 using Student's t-test for comparison between the cells treated with astaxanthin in Flag-XPC or pcDNA3 vector-transfected cells. (C) Cells were transfected with siRNA duplexes (200 nM) specific to XPC or scrambled (control) in complete medium prior to treatment with astaxanthin in complete medium for 24 h. The total RNA was isolated and subjected to real-time PCR. (D and E) After the above-mentioned treatment, cytotoxicity was determined by MTS and trypan blue exclusion assay. (F) After the cells were transfected with si-XPC or si-scrambled RNA, the cells were treated with astaxanthin (10 μM) for 24, 48, and 72 h, after which living cells were determined by trypan blue exclusion assay. The results (mean ± SEM) were from three independent experiments. **p < 0.01 using Student's t-test for comparison between the cells treated with astaxanthin in si-XPC RNA or si-scrambled RNA-transfected cells. (G) Left panel, A549 or H1975 cells were transfected with p38 MAPK siRNA and then co-treated with astaxanthin for 24 h. Right panel, cells were pretreated with SB203580 (5 μM) for 1 h and then co-treated with various concentration of astaxanthin for 24 h. Cytotoxicity was determined by MTS assay. **p < 0.01 using Student's t-test for comparison between the cells pretreated with or without p38 MAPK siRNA/SB203580 in astaxanthin exposed cells. (H) Cells were treated with astaxanthin (10 μM) and/or SB203580 (10 μM) for 1–3 days after which living cells were determined by MTS assay. **p < 0.01 using Student's t-test for comparison between cells treated with astaxanthin alone or with an astaxanthin and SB203580 combination.
Blocking p38 MAPK activation decreased astaxanthin-induced cytotoxicity and growth inhibition
In Fig. 4G, co-treatment with SB203580 significantly enhanced cell viability in astaxanthin-exposed A549 or H1975 cells, compared with astaxanthin treatment alone. SB203580 could decline growth inhibition effect after treatment with astaxanthin. Blocking p38 MAPK activity by SB203580 could less effectively inhibit cell growth than either drug alone after astaxanthin treatment (Fig. 4H). Taken together, activation of the p38 MAPK and down-regulation of XPC expression involved in astaxanthin-induced cytotoxicity and growth inhibition in NSCLC cells.
Combination treatment with astaxanthin enhanced the cytotoxic effect and growth inhibition of erlotinib
We attempted to determine whether astaxanthin could enhance the cytotoxic effect of erlotinib through down-regulating XPC expression in NSCLC cells. The effect of combined treatment on cell viability was examined by MTS assay. Combined treatment with astaxanthin and erlotinib for 24 h resulted in a greater loss of cell viability in A549 and H1975 cells than treatment with either erlotinib or astaxanthin alone (Fig. 5A). The CI values for astaxanthin and erlotinib were <1, indicating the combined treatment had a synergistic effect (Fig. 5B). In addition, A549 and H1975 cells were exposed to astaxanthin and/or erlotinib, and cell proliferation was determined 1–3 days after exposure. Astaxanthin and erlotinib co-treatment had a greater cell growth inhibition effect than either treatment alone (Fig. 5C). Next, the colony-forming assay was carried out to investigate whether astaxanthin and/or erlotinib affected long-term clonogenic cell survival. In Fig. 5D, the colony-forming ability of combined astaxanthin–erlotinib treatment was lower than that of erlotinib treatment alone in A549 (47.87% vs. 75.55%) and H1975 cells (41.35% vs. 79.05%). The result showed that combined astaxanthin and erlotinib had a synergistic cytotoxic effect on human NSCLC cells.
Fig. 5. Astaxanthin co-treatment with erlotinib synergistically enhanced cytotoxicity. (A) Erlotinib and astaxanthin were combined at a ratio of 1 : 2 and the MTS assay was used to analyze cell viability. (B) The mean CI values at a fraction affected (FA) of 0.50, 0.75, 0.90 for erlotinib and astaxanthin combined treatment were averaged for three independent experiments. (C) Cells were treated with astaxanthin (5 μM) and/or erlotinib (2.5, 5 μM) for 1–3 days after which living cells were determined by MTS assay. **p < 0.01 using Student's t-test for comparison between cells treated with a drug alone or with a astaxanthin/erlotinib combination. (D) Cells were treated with astaxanthin (2.5 μM) and/or erlotinib (1.25 μM) for 24 h, and then the colony forming ability of cells were determined by CFA assay. **p < 0.01 using Student's t-test for comparison between cells treated with a drug alone or with an astaxanthin/erlotinib combination. (E) A549 or H1975 cells were exposed to astaxanthin (5, 10, 20 μM) and erlotinib (10 μM) for 24 h. After treatment, total RNA was isolated and subjected to real-time PCR. The means ± standard deviation (SD) from four independent experiments. ** denotes p < 0.01, respectively, using Student's t-test for comparison between the cells treated with astaxanthin/erlotinib alone or combined. (F) After treatment as the above, cell extracts were examined by western blot analysis.
Astaxanthin down-regulated XPC protein and the mRNA level in erlotinib-treated human lung cancer cells
In order to assess the mechanism of the synergistic effect, we hypothesized that astaxanthin would affect XPC expression in erlotinib-treated NSCLC cells. To test this hypothesis, A549 and H1975 cells were exposed to astaxanthin for 24 h. In Fig. 5E and F, astaxanthin decreased XPC mRNA and protein levels in erlotinib-treated A549 and H1975 cells. Moreover, astaxanthin increased the phospho-p38 MAPK protein levels in erlotinib-treated NSCLC cells (Fig. 5F).
Transfection with MKK6E vectors decreased the XPC protein level as well as the cell survival by erlotinib and astaxanthin
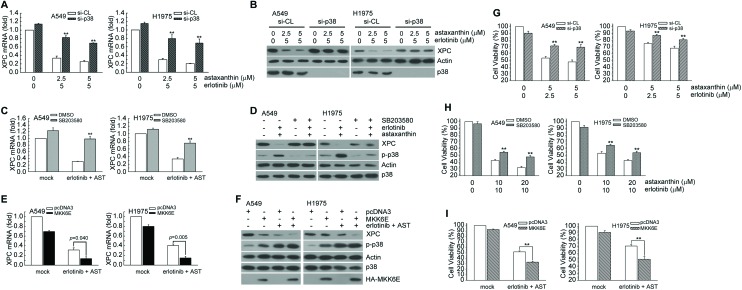
We investigated whether astaxanthin and erlotinib combination-mediated XPC down-regulation was correlated with p38 MAPK activation in NSCLC cells. In Fig. 6A–D, inactivation of p38 MAPK activity by knockdown p38 MAPK expression or pretreatment with SB203580 rescued XPC mRNA and protein levels in astaxanthin and erlotinib cotreated A549 and H1975 cells. In contrast, enforced MKK6E vector expression could further decrease XPC mRNA and protein expression (Fig. 6E and F). Inactivation of p38 MAPK activity by p38 MAPK siRNA transfection or pretreatment with specific p38 MAPK inhibitor recused viability after being decreased by astaxanthin and erlotinib (Fig. 6G and H); however, enforced expression of the MKK6E vector could further decrease A549 and H1975 cell viability (Fig. 6I). Taken together, astaxanthin enhanced erlotinib-induced cytotoxicity by p38 MAPK mediated XPC down-regulation in A549 and H1975 cells.
Fig. 6. Inactivation of p38 MAPK activity restored the suppressed XPC protein expression and cell survival in astaxanthin and erlotinib-exposed A549 and H1975 cells. (A and B) After cells were transfected with si-p38 MAPK RNA or scrambled (control), the cells were treated with astaxanthin and erlotinib in complete medium for 24 h. The results (mean ± SEM) were from 3 independent experiments. **p < 0.01, using Student's t-test for comparison between cells treated with astaxanthin and erlotinib in si-p38 MAPK RNA or si-scrambled RNA-transfected cells. The total RNA was isolated and subjected to real-time PCR and cell extracts were examined by western blot analysis. (C) Cells were pretreated with SB203580 (5 μM) for 1 h and then co-treated with astaxanthin (5 μM) and erlotinib (10 μM) for 24 h. Total RNA was isolated and subjected to real-time PCR. **p < 0.01 using Student's t-test for comparison between the cells treated with astaxanthin/erlotinib–DMSO or a combination of astaxanthin/erlotinib–SB203580. (D) After treatment of the above, the whole-cell extracts were collected for western blot analysis. (E) After MKK6E (3 μg) or pcDNA3 (3 μg) expression plasmids transfected into cells, the cells were treated with astaxanthin (5 μM) and erlotinib (5 μM) for an additional 24 h, and total RNA was isolated and subjected to real-time PCR. The means ± standard deviation (SD) from four independent experiments. ** denotes p < 0.01, using Student's t-test to compare cells treated with astaxanthin and erlotinib in MKK6E vs. pcDNA3-transfected cells. (F) After treatment as the above, whole-cell extracts were collected for western blot analysis. (G) After treatment as the (A). Cytotoxicity was determined by assessment with the MTS assay. *p < 0.05, **p < 0.01 by Student's t-test to compare cells treated with astaxanthin and erlotinib in si-p38 vs. scrambled (control) RNA-transfected A549 or H1975 cells. (H) After treatment as the (C). Cytotoxicity was determined by the MTS assay. **p < 0.01 using Student's t-test for comparison between the cells pretreated with or without SB203580 in astaxanthin and erlotinib-exposed cells. (I) After treatment as the (E). Cytotoxicity was determined by the MTS assay. **p < 0.01 using Student's t-test for comparison between the cells treated with astaxanthin and erlotinib in MKK6E vs. pcDNA3-transfected cells.
Discussion
Astaxanthin is a red carotenoid pigment having antioxidant activity and is found in marine environments with algae and aquatic animals.30 Several studies reveal the beneficial effects of astaxanthin against various diseases in both in vivo and in vitro systems.31–33 Astaxanthin protects somatic and germ cells from genotoxin induced DNA damage.34,35 The previous study showed that the neuroprotective effects of astaxanthin on H2O2-mediated apoptotic cell death occured via modulation of p38 MAPK and MEK signaling pathways and decreased H2O2-mediated cell death.36 In addition, astaxanthin treatment impedes the progression of mammary tumors in mice.37 Furthermore, Kurihara et al. reported that astaxanthin treatment inhibits the promotion of cancer metastasis in mice.38 Lastly, a previous study demonstrated that astaxanthin inhibits the development of hamster buccal pouch carcinogenesis carcinomas by inducing intrinsic apoptosis via coordinate inaction of both the ERK1/2 and AKT signals.39 Our results showed that phospho-MKK3/6 and phospho-p38 MAPK were significantly increased in A549 and H1975 cells in a dose and time-dependent manner after astaxanthin treatment. Inhibition of p38 MAPK activity restored the cell viability decreased by astaxanthin. Therefore, astaxanthin induced cytotoxic effects in human lung cancer cells proceeds via MKK3/6-p38 MAPK activation in human lung cancer cells.
XPC, a DNA repair factor, plays a critical role in preventing carcinogenesis.40,41 PTEN loss suppresses expression of XPC through the AKT and p38 signaling in human HaCaT keratinocytes.42 Long-term silencing of the XPC gene expression led to an enhanced sensitivity to topoisomerase II inhibitor etoposide.43 We found that astaxanthin treatment decreased XPC expression in a p38 MAPK activation manner. Enforced expression of the Flag-XPC expression vector cloud rescued A549 and H1975 cell viability decreased by astaxanthin and reversed the growth inhibition effect. We also found that astaxanthin-mediated XPC down-regulation plays a role in enhancing the sensitivity of erlotinib in lung cancer. Reported by Sun et al., miR-346 directly targets 3′-UTR (3′-untranslated regions) of XPC mRNA which contributes to a decrease of XPC protein in A549 cells. The deficiency of XPC induces the accumulation of the endogenous DNA damage, and then activates the ERK/Snail/E-cadherin pathway.44 In our study, astaxanthin decreased XPC protein and mRNA levels through the enhancement of protein and mRNA instability of XPC by p38 MAPK activation. p38 MAPK activation decreased XPC protein level by triggering XPC protein degradation via the ubiquitin-mediated 26S proteasome pathway in astaxanthin-treated A549 and H1975 cells. Therefore, the p38 MAPK signal pathway plays a role in repression of XPC expression in astaxanthin-treated NSCLC cells.
Previous reports have concluded that upregulation of p38 MAPK signaling is responsible for the enhanced IL-8 secretion in the erlotinib-resistant tumor cells.45 Recently, a study indicated that the combination treatment of chelerythrine and erlotinib significantly decreased cell proliferation and reduced clonogenicity. The possible mechanism was the combination of the treatment being highly effective at blocking EGF induced phosphorylation of p38 MAPK, STAT3, ERK1/2, and BAD proteins.46 In our previous study, down-regulation of AKT-mediated thymidylate synthase expression by salinomycin enhanced the erlotinib-induced cytotoxicity in NSCLC cells.47 In this study, combined treatment with astaxanthin enhanced p38 MAPK activation and down-regulated the expression of XPC in erlotinib-exposed A549 and H1975 cells, and subsequently resulted in synergistic cytotoxic effects in NSCLC cells.
Taken together, we found astaxanthin enhanced NSCLC cells sensitivity to erlotinib as a result of MKK3/6-p38 MAPK mediated XPC decreases. These results may provide the rational design of future drug regimens incorporating astaxanthin and erlotinib for the treatment of NSCLC. Although further studies are required to evaluate the effect of astaxanthin and erlotinib combination in vivo, the concept of erlotinib combined with astaxanthin seems to be a strategy for the treatment of NSCLC.
Conflicts of interest
There are no conflicts of interest to declare.
Acknowledgments
This study was funded by grants from the Ministry of Science and Technology, Taiwan, Grant MOST 107-2320-B-415-007.
References
- Pfister D. G., Johnson D. H., Azzoli C. G., Sause W., Smith T. J., Baker, Jr. S., Olak J., Stover D., Strawn J. R., Turrisi A. T., Somerfield M. R. J. Clin. Oncol. 2004;22:330–353. doi: 10.1200/JCO.2004.09.053. [DOI] [PubMed] [Google Scholar]
- Chen W., Zheng R., Baade P. D., Zhang S., Zeng H., Bray F., Jemal A., Yu X. Q., He J. CA-Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. CA-Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Salomon D. S., Brandt R., Ciardiello F., Normanno N. Crit. Rev. Oncol. Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- Arteaga C. L. J. Clin. Oncol. 2003;21:289s–291s. doi: 10.1200/JCO.2003.10.523. [DOI] [PubMed] [Google Scholar]
- Raymond E., Faivre S., Armand J. P., Drugs, 2000, 60Suppl 1 , 15 –23 , ; discussion 41–12 . [DOI] [PubMed] [Google Scholar]
- Felip E., Rosell R. Drugs Today. 2006;42:147–156. doi: 10.1358/dot.2006.42.3.957358. [DOI] [PubMed] [Google Scholar]
- Kris M. G., Natale R. B., Herbst R. S., Lynch, Jr. T. J., Prager D., Belani C. P., Schiller J. H., Kelly K., Spiridonidis H., Sandler A., Albain K. S., Cella D., Wolf M. K., Averbuch S. D., Ochs J. J., Kay A. C. J. Am. Med. Assoc. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- Legerski R., Peterson C. Nature. 1992;359:70–73. doi: 10.1038/359070a0. [DOI] [PubMed] [Google Scholar]
- Sands A. T., Abuin A., Sanchez A., Conti C. J., Bradley A. Nature. 1995;377:162–165. doi: 10.1038/377162a0. [DOI] [PubMed] [Google Scholar]
- Cheo D. L., Meira L. B., Hammer R. E., Burns D. K., Doughty A. T., Friedberg E. C. Curr. Biol. 1996;6:1691–1694. doi: 10.1016/s0960-9822(02)70794-x. [DOI] [PubMed] [Google Scholar]
- Cheo D. L., Burns D. K., Meira L. B., Houle J. F., Friedberg E. C. Cancer Res. 1999;59:771–775. [PubMed] [Google Scholar]
- Rezvani H. R., Kim A. L., Rossignol R., Ali N., Daly M., Mahfouf W., Bellance N., Taieb A., de Verneuil H., Mazurier F., Bickers D. R. J. Clin. Invest. 2011;121:195–211. doi: 10.1172/JCI40087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T., Srivastava A. K., Han C., Yang L., Zhao R., Zou N., Qu M., Duan W., Zhang X., Wang Q. E. Oncotarget. 2015;6:10060–10072. doi: 10.18632/oncotarget.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. E., Han C., Zhang B., Sabapathy K., Wani A. A. Cancer Res. 2012;72:666–675. doi: 10.1158/0008-5472.CAN-11-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S., Kogure K., Abe K., Kimata Y., Kitahama K., Yamashita E., Terada H. Biochim. Biophys. Acta. 2001;1512:251–258. doi: 10.1016/s0005-2736(01)00326-1. [DOI] [PubMed] [Google Scholar]
- Higuera-Ciapara I., Felix-Valenzuela L., Goycoolea F. M. Crit. Rev. Food Sci. Nutr. 2006;46:185–196. doi: 10.1080/10408690590957188. [DOI] [PubMed] [Google Scholar]
- Campoio T. R., Oliveira F. A., Otton R. Toxicol. In Vitro. 2011;25:1448–1456. doi: 10.1016/j.tiv.2011.04.018. [DOI] [PubMed] [Google Scholar]
- Marin D. P., Bolin A. P., Macedo Rde C., Sampaio S. C., Otton R. Int. Immunopharmacol. 2011;11:103–109. doi: 10.1016/j.intimp.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Wolf A. M., Asoh S., Hiranuma H., Ohsawa I., Iio K., Satou A., Ishikura M., Ohta S. J. Nutr. Biochem. 2010;21:381–389. doi: 10.1016/j.jnutbio.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Lee D. H., Lee Y. J., Kwon K. H. J. Clin. Biochem. Nutr. 2010;47:121–129. doi: 10.3164/jcbn.10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao R., Nelson O. L., Park J. S., Mathison B. D., Thompson P. A., Chew B. P. Anticancer Res. 2010;30:2171–2175. [PubMed] [Google Scholar]
- Song X., Wang M., Zhang L., Zhang J., Wang X., Liu W., Gu X., Lv C. Toxicol. Mech. Methods. 2012;22:679–686. doi: 10.3109/15376516.2012.717119. [DOI] [PubMed] [Google Scholar]
- Macedo R. C., Bolin A. P., Marin D. P., Otton R. Eur. J. Nutr. 2010;49:447–457. doi: 10.1007/s00394-010-0103-1. [DOI] [PubMed] [Google Scholar]
- Lordan S., O'Neill C., O'Brien N. M. Br. J. Nutr. 2008;100:287–296. doi: 10.1017/S0007114507898643. [DOI] [PubMed] [Google Scholar]
- Nagendraprabhu P., Sudhandiran G. Invest. New Drugs. 2011;29:207–224. doi: 10.1007/s10637-009-9342-5. [DOI] [PubMed] [Google Scholar]
- Ko J. C., Ciou S. C., Jhan J. Y., Cheng C. M., Su Y. J., Chuang S. M., Lin S. T., Chang C. C., Lin Y. W. Mol. Cancer Res. 2009;7:1378–1389. doi: 10.1158/1541-7786.MCR-09-0051. [DOI] [PubMed] [Google Scholar]
- Tsai M. S., Weng S. H., Chen H. J., Chiu Y. F., Huang Y. C., Tseng S. C., Kuo Y. H., Lin Y. W. Mol. Cancer Ther. 2012;11:561–571. doi: 10.1158/1535-7163.MCT-11-0684. [DOI] [PubMed] [Google Scholar]
- Peters G. J., van der Wilt C. L., van Moorsel C. J., Kroep J. R., Bergman A. M., Ackland S. P. Pharmacol. Ther. 2000;87:227–253. doi: 10.1016/s0163-7258(00)00086-3. [DOI] [PubMed] [Google Scholar]
- Lai J. P., Jiang Y., He X. W., Huang J. C., Chen F. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2004;804:25–30. doi: 10.1016/j.jchromb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Bennedsen M., Wang X., Willen R., Wadstrom T., Andersen L. P. Immunol. Lett. 1999;70:185–189. doi: 10.1016/s0165-2478(99)00145-5. [DOI] [PubMed] [Google Scholar]
- Hussein G., Goto H., Oda S., Sankawa U., Matsumoto K., Watanabe H. Biol. Pharm. Bull. 2006;29:684–688. doi: 10.1248/bpb.29.684. [DOI] [PubMed] [Google Scholar]
- Kamath B. S., Srikanta B. M., Dharmesh S. M., Sarada R., Ravishankar G. A. Eur. J. Pharmacol. 2008;590:387–395. doi: 10.1016/j.ejphar.2008.06.042. [DOI] [PubMed] [Google Scholar]
- Tripathi D. N., Jena G. B. Toxicology. 2008;248:96–103. doi: 10.1016/j.tox.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Tripathi D. N., Jena G. B. Chem.-Biol. Interact. 2009;180:398–406. doi: 10.1016/j.cbi.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Choi W., Lee J. H., Jeon S. J., Choi Y. H., Kim B. W., Chang H. I., Nam S. W. J. Microbiol. Biotechnol. 2009;19:1355–1363. doi: 10.4014/jmb.0906.06003. [DOI] [PubMed] [Google Scholar]
- Chew B. P., Park J. S., Wong M. W., Wong T. S. Anticancer Res. 1999;19:1849–1853. [PubMed] [Google Scholar]
- Kurihara H., Koda H., Asami S., Kiso Y., Tanaka T. Life Sci. 2002;70:2509–2520. doi: 10.1016/s0024-3205(02)01522-9. [DOI] [PubMed] [Google Scholar]
- Kavitha K., Kowshik J., Kishore T. K., Baba A. B., Nagini S. Biochim. Biophys. Acta. 2013;1830:4433–4444. doi: 10.1016/j.bbagen.2013.05.032. [DOI] [PubMed] [Google Scholar]
- Hollander M. C., Philburn R. T., Patterson A. D., Velasco-Miguel S., Friedberg E. C., Linnoila R. I., Fornace, Jr. A. J. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13200–13205. doi: 10.1073/pnas.0503133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W., Soltani K., Ming M., He Y. Y. Cancer Prev. Res. 2012;5:1155–1162. doi: 10.1158/1940-6207.CAPR-12-0185-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming M., Feng L., Shea C. R., Soltani K., Zhao B., Han W., Smart R. C., Trempus C. S., He Y. Y. Cancer Res. 2011;71:5287–5295. doi: 10.1158/0008-5472.CAN-10-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despras E., Pfeiffer P., Salles B., Calsou P., Kuhfittig-Kulle S., Angulo J. F., Biard D. S. Cancer Res. 2007;67:2526–2534. doi: 10.1158/0008-5472.CAN-06-3371. [DOI] [PubMed] [Google Scholar]
- Sun C. C., Li S. J., Yuan Z. P., Li D. J. Aging. 2016;8:2509–2524. doi: 10.18632/aging.101080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fernando R. I., Hamilton D. H., Dominguez C., David J. M., McCampbell K. K., Palena C. Oncotarget. 2016;7:42031–42044. doi: 10.18632/oncotarget.9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Yang Z., Zhang L., Song C., Li Y., Zhang X. PLoS One. 2017;12:e0175466. doi: 10.1371/journal.pone.0175466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C. L., Chen J. C., Wu C. H., Peng Y. S., Chen W. C., Zheng H. Y., Jian Y. J., Wei C. L., Cheng Y. T., Lin Y. W. Exp. Cell Res. 2017;357:59–66. doi: 10.1016/j.yexcr.2017.04.026. [DOI] [PubMed] [Google Scholar]