Figure 1.
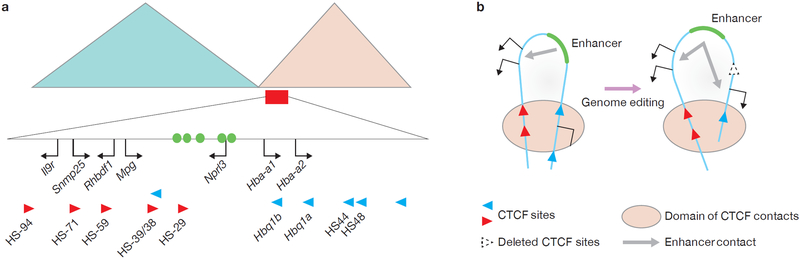
The α-globin cluster in mouse ESCs and primary erythroid cells. (a) The large triangles represent Hi-C results showing two adjacent TADs in mouse ESCs with a boundary between them1. The domain within the red rectangle includes the α-globin genes and their enhancers (green circles), and upstream widely expressed genes. Red and blue arrowheads denote DNase I hypersensitive sites (HSs) bound by CTCF. The directionality of the arrowhead indicates the orientation of the underlying CTCF-binding motif. (b) A simplified model depicting interaction of convergently arranged CTCF sites to form a ‘hairpin’ structure described by Hanssen et al.9. The head of the pin is a loop that encloses an enhancer and its target genes but excludes an unrelated gene. When genome editing is used to remove the CTCF site at one end of the loop, the loop still forms but is enlarged by interaction of the next appropriate CTCF site. This results in incorporation of a new gene that then becomes a target of the enhancer.