Bioinformatics, (2018) https://doi.org/10.1093/bioinformatics/bty357
The author wishes to apologize for a mistake in Figure 2 in the above manuscript. The figure appears correctly below:
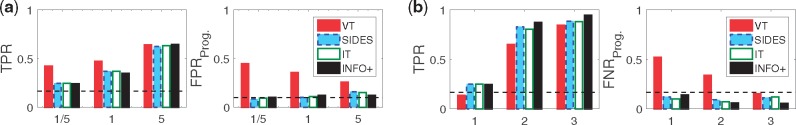
Fig. 2.
When biomarkers have both prognostic/predictive strength (M-1) VT achieves higher TPR, otherwise (M-2) the gains in TPR are vanishing. In terms of , VT always has very high error rate on selecting solely prognostic biomarkers as predictive, and it performs worse than random selection. This is the average TPR/ over 200 simulated datasets for three different values of the predictive strength θ: 1/5 means a strongly prognostic signal, 1 means equal strength between prognostic and predictive signals, and 5 means a strongly predictive signal. The sample size is 2000, and the dimensionality p = 30 biomarkers. Dashed lines show the TPR/ if we were ranking the biomarkers at random. (a) M-1: Biomarkers can be both prognostic and predictive. (b) M-2: Biomarkers are solely either prognostic or predictive
The paper has been corrected online.