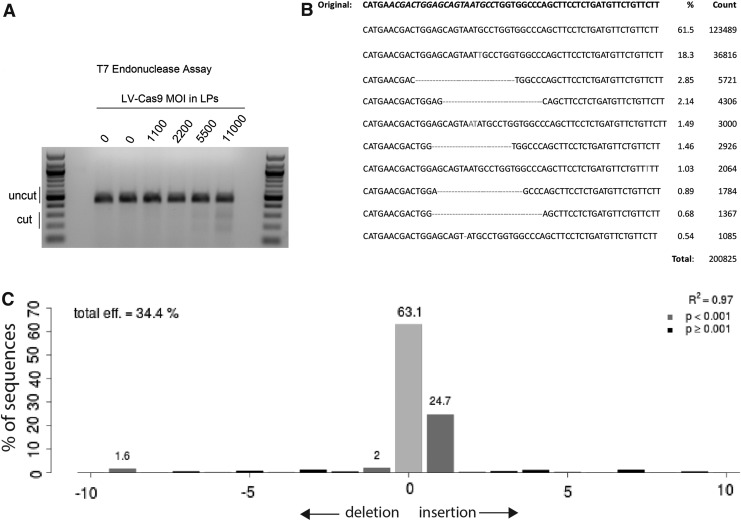
Figure 2.
LV-Cas9-mediated gene editing at the Fah−/− locus in fibroblasts. (A) A 452 base pair (bp) region around the HT1 single nucleotide polymorphism was amplified using polymerase chain reaction (PCR), re-annealed, and analyzed using a T7 endonuclease assay, which cleaves DNA at mismatched sequences. The lower bands in the gel indicated that LV-Cas9 did disrupt the Fah locus. (B) Gene disruption was confirmed by next-generation sequencing of amplicons. The top line is the non-modified mutant sequence, and the lines below indicate the frequency of each modified sequence at this locus. The guide sequence is italicized in the top line. (C) Representative Tracking of Indels by DEcomposition analysis of cells treated with LV-Cas9 at a multiplicity of infection of 8,000 lentiviral (LV) particles. The Cas9 efficiency in this sample was 34.4%, with the most frequent indel being a +1 insertion (24.7%).