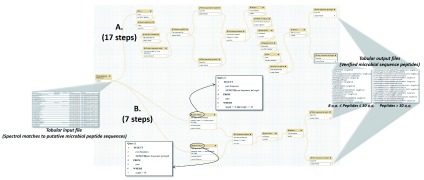
Figure 3. A metaproteomics workflow using Query Tabular.
This workflow verifies the presence of detected microbial peptides by matching peptides against the NCBI nr protein sequence database using the BLASTP tool. ( A) Using conventional Galaxy text manipulation tools, the workflow requires 17 steps to achieve desired outputs. ( B) When utilizing Query Tabular, desired results are obtained in seven steps. The figure also displays the structure of the tabular input file containing peptide sequences putatively from microbes in the sample, and the tabular output file which contains verified microbial peptide sequences. One output contains peptides between 8 and 30 amino acids (a.a.) in length, while the other contains peptides 30 a.a. or greater in length, due to BLASTP requiring separate analysis based on length of peptide sequences. The two inset boxes show the two SQL queries used in this workflow, made possible by Query Tabular.