Abstract
Background
Chronic lymphocytic leukemia (CLL) has a heterogeneous clinical course. Beside patients requiring immediate treatment, others show an initial indolent phase followed by progression and others do not progress for decades. The latter two subgroups usually display mutated IGHV genes and a favorable FISH profile.
Patients and methods
Patients with absence of disease progression for over 10 years (10–34) from diagnosis were defined as ultra-stable CLL (US-CLL). Forty US-CLL underwent extensive characterization including whole exome sequencing (WES), ultra-deep sequencing and copy number aberration (CNA) analysis to define their unexplored genetic landscape. Microarray analysis, comparing US-CLL with non-US-CLL with similar immunogenetic features (mutated IGHV/favorable FISH), was also carried out to recognize US-CLL at diagnosis.
Results
WES was carried out in 20 US-CLL and 84 non-silent somatic mutations in 78 genes were found. When re-tested in a validation cohort of 20 further US-CLL, no recurrent lesion was identified. No clonal mutations of NOTCH1, BIRC3, SF3B1 and TP53 were found, including ATM and other potential progression driving mutations. CNA analysis identified 31 lesions, none with known poor prognostic impact. No novel recurrent lesion was identified: most cases showed no lesions (38%) or an isolated del(13q) (31%). The expression of 6 genes, selected from a gene expression profile analysis by microarray and quantified by droplet digital PCR on a cohort of 79 CLL (58 US-CLL and 21 non-US-CLL), allowed to build a decision-tree capable of recognizing at diagnosis US-CLL patients.
Conclusions
The genetic landscape of US-CLL is characterized by the absence of known unfavorable driver mutations/CNA and of novel recurrent genetic lesions. Among CLL patients with favorable immunogenetics, a decision-tree based on the expression of 6 genes may identify at diagnosis patients who are likely to maintain an indolent disease for decades.
Keywords: chronic lymphocytic leukemia, ultra-stable disease, whole exome sequencing, copy number aberrations, gene expression profile
Key Message
The genetic landscape of ultra-stable CLL is characterized by a marked genetic stability with a low mutation and CNA load, and the absence of unfavorable or recurrent genetic lesions. Among CLL with favorable immunogenetic features, ultra-stable patients can be identified at diagnosis by a decision-tree model based on the expression of 6 genes.
Introduction
Chronic lymphocytic leukemia (CLL) shows an extremely heterogeneous clinical course, linked to immunogenetic markers with well-known prognostic implications [1–3]. Beside cases with aggressive disease at onset often requiring immediate treatment (unmutated IGHV genes, ATM/TP53 disruption), there are patients with an initial indolent phase followed by disease progression and others who do not progress for decades or ever, both usually characterized by mutated IGHV genes and a favorable FISH profile.
Next generation sequencing (NGS) technologies have identified previously unknown genetic lesions mainly affecting NOTCH1, SF3B1, MYD88 and BIRC3 genes [4]; their integration with FISH abnormalities has further improved the prognostic stratification of CLL patients [5]. More recently, the presence of TP53 mutated subclones in untreated CLL has been associated to the same poor prognostic impact of clonal TP53 lesions [6]. Furthermore, data arising from genome-wide copy number aberration (CNA) studies have identified additional genetic lesions playing a role on CLL clinical outcome [7, 8].
We have previously reported on the distinctive biologic profile of patients with ultra-stable disease (US-CLL) for more than 10 years from diagnosis [9, 10]. Here, we have applied whole exome sequencing (WES), ultra-deep sequencing and CNA analysis to 40 US-CLL cases to further investigate the genetic landscape of this specific subgroup. A microarray analysis was carried out to identify a gene signature capable of recognizing US-CLL patients at diagnosis among cases with a favorable immunogenetic profile.
Patients and methods
Study population
US-CLL was defined as follows: absence of treatment requirement for at least 10 years from diagnosis, no change in clinical stage, no clinical signs of disease activity and regardless of the lymphocyte count [9]. The discovery cohort for WES analysis consisted of 20 US-CLL patients. A second cohort of 20 US-CLL patients was used to screen gene mutations identified by WES (screening cohort). Overall, the median follow-up from diagnosis was 15 years (10–34; supplementary Table S1, available at Annals of Oncology online). CD38 and ZAP70 expression, FISH and IGHV gene analyses were carried out as described [11, 12]. All samples satisfied the diagnostic criteria for CLL, including a CLL lymphocyte count >5.0 ×109/L at diagnosis [13].
In the screening cohort, TP53 (exons 4–9), SF3B1 (exons 14–16), NOTCH1 (exon 34), BIRC3 (exons 6–9) were analyzed by DNA direct sequencing (ABI PRISM 3100 Genetic Analyzer, Applied Biosystems) [2, 14–16].
All patients provided their informed consent to blood and germline material collection, and subsequent biological analyses in accordance with the Declaration of Helsinki. This study was approved by the Ethical Committee (2182/16.06.2011).
WES, identification of tumor-specific variants, validation and screening of mutated genes
Genomic DNA (gDNA) from mononuclear peripheral blood cells of 20 US-CLL, including paired germline DNA (saliva) in 14, was used for WES analysis on Illumina HiSeq 2000 analyzer (Illumina, San Diego, CA; supplementary Tables S2 and S3, available at Annals of Oncology online).
The most frequent candidate non-silent somatic mutations (≥2 cases) identified by WES were subjected to validation in the discovery cohort by Sanger sequencing on tumor and germline gDNA. The recurrently mutated genes (≥2 cases) were sequenced by Sanger in the screening cohort (supplementary Material, available at Annals of Oncology online).
Ultra-deep NGS of TP53 gene
Exons 4–9 including splicing sites of the TP53 gene underwent an ultra-deep NGS approach on the Genome Sequencer Junior instrument (Roche-454) (Roche, Mannheim, Germany) in 35 US-CLL (19 from the discovery cohort; 16 from the screening cohort; supplementary Table S2, available at Annals of Oncology online) [6].
CNA analysis by high-density Cytoscan array
Genome-wide DNA profiles were obtained from gDNA of 29 US-CLL patients (16 from the discovery cohort; 13 from the screening cohort) using the Affymetrix Cytoscan high-density (HD) Array and standard protocols (Affymetrix, Santa Clara, CA; supplementary Table S2, available at Annals of Oncology online).
Gene expression signature by microarray and droplet digital PCR
Twelve US-CLL were studied by oligonucleotide arrays (GeneChip® Human Genome U133 Plus 2.0 Affymetrix; supplementary Table S2, available at Annals of Oncology online) [17]. They were compared with 12 non-US-CLL with mutated IGHV and favorable FISH lesions, evaluated both at diagnosis (T1) and at first progression (T2) (occurred after a median of 3 years from diagnosis; range 1–5), in order to select the genes that most significantly identified US-CLL (supplementary Figure S1, available at Annals of Oncology online).
The expression of the selected genes was validated using the QX200™ droplet digital PCR (ddPCR) (Bio-Rad, Hercules, CA) on 12 US-CLL and 12 non-US-CLL at T1 (supplementary Table S2, available at Annals of Oncology online) and thereafter evaluated on an independent cohort of 55 CLL cases, for a total of 79 cases (58 US-CLL; 21 non-US-CLL; supplementary Table S4, available at Annals of Oncology online). The gene expression data were used to build a decision-tree capable of identifying US-CLL patients at diagnosis (supplementary Material, available at Annals of Oncology online).
The different numbers of cases tested for the different methods (supplementary Table S2, available at Annals of Oncology online) was due to sample availability.
Results
Conventional biologic features
All US-CLL cases were IGHV mutated, with the IGHV3-30 gene being the most frequent (7/40, 17.5%). No recurrent stereotyped B-cell receptor (BCR) was identified. Six/40 (15%) US-CLL showed a stereotyped BCR: subset #4 (both IGHV4-34) in 2, subset #202 in 1, subset #19, #67 and #90 in 3 cases, respectively (all associated with the VH3 family). There was no difference with the frequency of stereotyped BCR in non-US-CLL (4/17, 23.5%, subsets #1, #4, #202 and #95) reported in the manuscript (P = 0.46) and with the non-US CLL with mutated IGHV and favorable FISH of our database (17/116, 14.6%; P = 1; data not shown).
FISH results, available in 26 cases, showed del(13q) in 19/26 (73%) and no lesions in 7 (27%). CD38 and ZAP70 proved negative in 37/37 and 22/24 evaluated cases, respectively (supplementary Table S1, available at Annals of Oncology online).
WES analysis of US-CLL in the discovery cohort
WES analysis of the 14 cases with paired germline DNA identified 84 non-silent somatic mutations in 78 genes. The mutation load was 6 mutations/case (range 1–12); only 1 case showed >10 mutations. Mutations were predominantly missense substitutions (88%) and infrequently frameshift/in frame deletions (6%) or nonsense mutations (6%). All 78 genes are annotated in the COSMIC database. The PolyPhen-2 algorithm predicted as probably or possibly damaging 65% of mutations, while the remaining 35% were predicted as benign (supplementary Table S5, available at Annals of Oncology online).
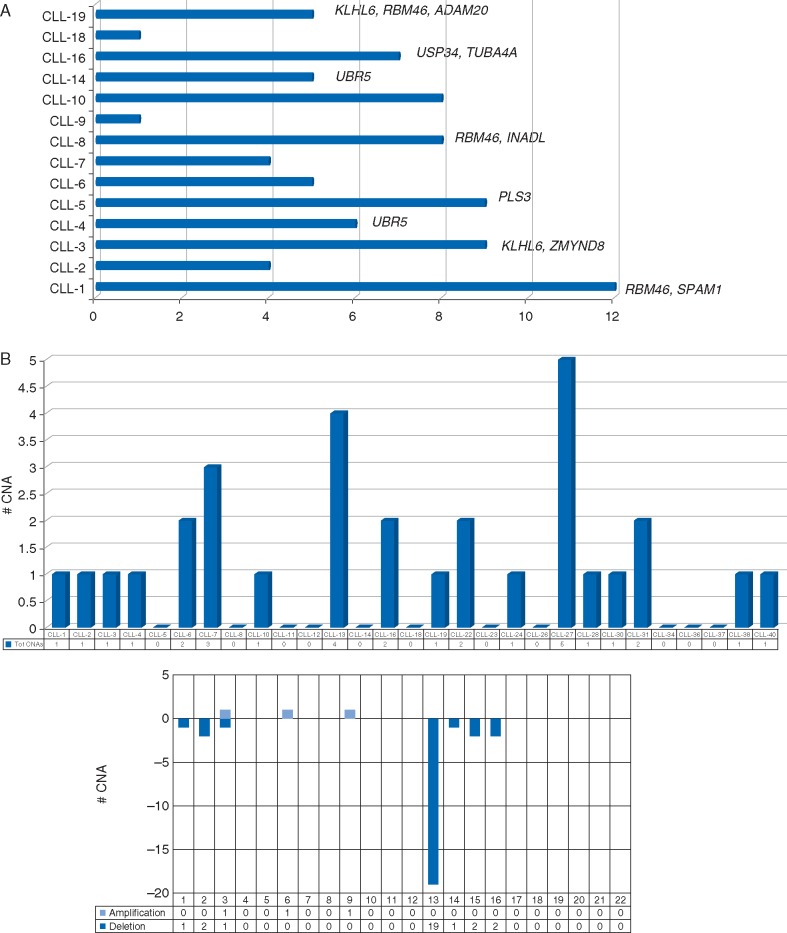
The remaining 6 cases without germline DNA evaluated by WES were pooled with the other 14 to assess the recurrence of mutated genes, after the exclusion of SNPs (dbSNP). Twelve genes were considered recurrent (mutated in ≥2 cases): ADAM20, INADL, KLHL6, PLS3, PRDM9, PTPRT, SPAM1, RBM46, TUBA4A, UBR5, USP34 and ZMYND8. Sanger sequencing validated 92% of mutations in 10 genes (ADAM20, INADL, KLHL6, PLS3, SPAM1, RBM46, TUBA4A, UBR5, USP34 and ZMYND8) with RBM46 somatically mutated in 3/20 patients (15%) (Figure 1A; supplementary Table S6, available at Annals of Oncology online).
Figure 1.
(A) WES in the discovery cohort. Number of non-silent somatic mutations per case. The 10 genes recurrent in ≥2 cases are reported. (B) CNA analysis. Number of CNA per case and per chromosome.
WES analysis of TP53, ATM, NOTCH1, SF3B1, BIRC3 and MYD88 proved wild-type in all cases; a FBXW7 somatic mutation (c.2171G>A, p.G305R) was identified in 1 case.
Most samples (13/20, 65%) evaluated for WES were collected after 10 or more years from diagnosis (see section ‘Discussion’).
Analysis of recurrently mutated genes in the screening cohort
Of the 10 recurrent genes, we excluded those already reported in CLL across different prognostic subgroups, mostly aggressive, therefore unlikely to bear prognostic/pathogenetic significance in a cohort of US-CLL (see section ‘Discussion’). We thus analyzed the whole codifying region and consensus splice sites of the ADAM20, SPAM1, TUBA4 and RBM46 genes in 20 further US-CLL. No additional mutated case was identified. Sanger sequencing of TP53, NOTCH1, SF3B1 and BIRC3 resulted wild-type in all cases.
Ultra-deep NGS of TP53 gene
Bioinformatic analysis revealed TP53 subclonal mutations in 2/35 (5.7%) US-CLL. Patient CLL-16 showed two missense and two nonsense subclonal mutations, with a median variant allele frequency (VAF) corrected for tumor representation (see supplementary Material, available at Annals of Oncology online) of 2.15% (range 1.3–2.3), while patient CLL-27 showed a single subclonal splice site mutation with a VAF of 14.6% (supplementary Table S7, available at Annals of Oncology online). All subclonal mutations were confirmed in an independent ultra-deep experiment and by AS-PCR. The location and in silico analysis of these mutations (IARC TP53 database, Polyphen algorithm) suggest that they may be deleterious.
The clinical history and follow-up of these two patients were divergent (see supplementary Material, available at Annals of Oncology online).
CNA analysis
CNA analysis identified 31 lesions (90% losses, 10% gains) in 29 cases, giving a CNA load of 1 lesion/case (range 0–5); only 3 cases showed ≥3 lesions (Figure 1B; supplementary Table S8, available at Annals of Oncology online). Eleven cases (38%) showed no lesions, 15 cases (52%) del(13q)—isolated in 10 cases and with additional non-canonical CNAs in 5—and 3 cases (10%) showed 1, 1 and 2 lesions, respectively. Regarding the size, del(13q) included the RB1 gene in 8/15 cases; of the 12 additional CNA, all were <5 Mbp (below the conventional cytogenetic sensitivity) and 3 were focal with only 1 gene involved.
No recurrent CNA was found, apart from del(13q), and no CNA with known poor prognostic impact [8]. None of the 78 genes mutated at WES were concomitantly affected by CNAs and no association between the 10 recurrent mutated genes and CNAs was found.
More than half of the samples (16/29, 55%) evaluated for CNAs were collected after 10 or more years from diagnosis (see section ‘Discussion’). In order to exclude the effect of clonal evolution, we carried out longitudinal CNA experiments in four US-CLL cases with del(13q) (5–8 years interval between two samples). In all cases, we observed the absence of any additional chromosomal lesion acquired over time, the presence of del(13q) only and, in two patients, an identical start-end of deletion 13q after 6 and 8 years, respectively, a further measure of their stability (data not shown).
Gene expression signature and development of the decision-tree for US-CLL identification
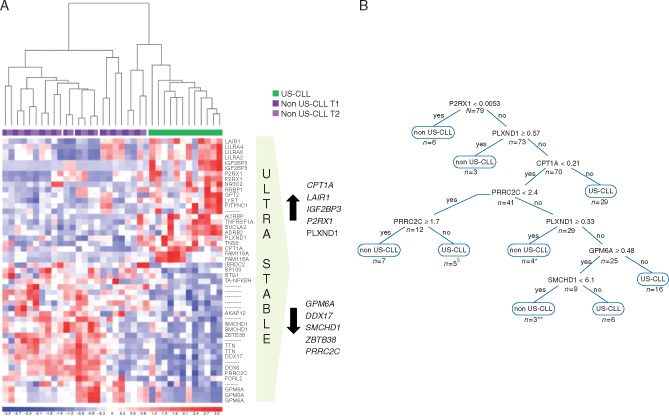
To identify genes characterizing US-CLL cases, two t-tests were carried out: US versus non-US-CLL at T1 (116 probe sets) and US versus non-US-CLL at T2 (635 probe sets). The two analyses were matched and resulted in a list of 32 selected genes that most significantly differentiated US-CLL from non-US-CLL, remaining unchanged between T1 and T2 (Figure 2A; supplementary Table S9, available at Annals of Oncology online). Ten were the most informative genes—GPM6A, DDX17, SMCHD1, ZBTB38, PRRC2C (downmodulated in US-CLL) and CPT1A, LAIR1, IGF2BP3, P2RX1, PLXND1 (upmodulated in US-CLL)—according to statistical and functional relevance (supplementary Table S10, available at Annals of Oncology online).
Figure 2.
(A) Gene expression profile analysis. The figure shows the 32 genes that most significantly identify US-CLL patients (see supplementary Figure S1, available at Annals of Oncology online for more details on microarray work-flow) at the supervised analysis. On the right, the 10 candidate classifier genes. (B) Decision-tree. The decision-tree is derived from the best predictive model in the R output, identifying eight subgroups (nodes) and six associated factors. The final decision-tree had the first split at P2RX1 expression value of 0.0053, the second decision node at PLXND1 expression value of 0.57, the third at CPT1A expression value of 0.21. In the fourth split for PRRCR2 expression values between 1.7 and 2.4 the patient was classified as US-CLL, for values <1.7 as non-US-CLL, for values ≥2.4 the evaluation of the next genes was required. The expression values derive from ddPCR quantification and represent an absolute measure of copies of each target gene/μl of reaction. §One non-US-CLL misclassified as US-CLL. *Two US-CLL misclassified as non-US-CLL. **One US-CLL misclassified as non-US-CLL.
The validation of the 10 genes using the ddPCR platform was first conducted on the microarray cohort (12 US-CLL and 12 non-US-CLL) which confirmed the downmodulation of GPM6A (P = 0.021), SMCHD1 (P = 0.039) and PRRC2C (P = 0.05) and the upmodulation of CPT1A (P = 0.002), LAIR1 (P = 0.022), IGF2BP3 (P = 0.044), P2RX1 (P = 0.0001) and PLXND1 (P = 0.039) in US-CLL, with concordance between microarray and ddPCR (supplementary Figure S2, available at Annals of Oncology online). The ddPCR analysis of DDX17 and ZBTB38 did not reach statistical significance. We thus analyzed by ddPCR the expression of the resulting 8 genes (GPM6A, SMCHD1, PRRC2C, CPT1A, LAIR1, IGF2BP3, P2RX1, PLXND1) on an independent cohort of 55 CLL patients. To build a prediction model capable of identifying US-CLL at diagnosis, the ddPCR results from the two cohorts were pooled. A decision-tree was generated to subdivide patients most at risk of being US-CLL or non-US-CLL according to the expression of the eight genes (associated factors). The final decision-tree analysis identified eight nodes and six associated factors (P2RX1, PLXND1, CPT1A, PRRC2C, GPM6A, SMCHD1; Figure 2B). To minimize the bias associated with overfitting, a 10-fold cross-validation was used; the dataset was divided into 10 parts: 9 were used for training and 1 for testing; the process was then repeated until all parts were tested. Only four patients were misclassified by the model, three US-CLL and one non-US-CLL. Thus, the classification tree showed a high specificity (95.2%), sensitivity (94.8%) and accuracy (94.9%), indicative of an appropriate and clinically meaningful screening tool.
Discussion
The interest in characterizing US-CLLs derives from the need of elucidating the bases of CLL stability/progression. In the present study, we investigated the genetic landscape of US-CLL patients, to recognize early cases with a high probability of not progressing for over a decade.
This report extends our previous studies [9, 18]. The first described a distinct biologic profile—negative CD38, mutated IGHV genes, absence of del(11q) and TP53 deletion/mutations—that could identify patients with a very favorable prognostic likelihood [9]. The second showed that CLL cases undergoing spontaneous clinical regression, the furthest expression of a benign clinical course, were associated with a distinctive gene profile including the expression of BCR signaling-related genes [18].
Along this line, the present series consists of US-CLL patients with a median follow-up from diagnosis of 15 years (range 10–34), all showing mutated IGHV genes, absence of unfavorable FISH abnormalities and of CD38/ZAP70 expression.
WES identified six mutations/case on average, a significantly lower load compared with that documented in other CLL series and in our previous study on chemo-refractory CLL [19–23]. No recurrently mutated gene could be associated with US-CLL. RBM46—that codifies for a RNA binding motif protein—recurred in 3/40 cases (7.5%), but its biologic relevance remains elusive. Occasionally reported in other hematologic diseases [24, 25], it has never been reported in CLL apart from a single case with a silent mutation [26].
Regarding the remaining nine gene mutations identified in two cases each, some have already been reported in CLL but across different prognostic subgroups (i.e. CLL with unmutated IGHV or TP53 disruption or progressive/chemorefractory disease [19–23, 26–28]); also KLHL6 mutations, known to be associated with mutated IGHV CLL, do not discriminate the indolent ones, since they have been reported also in patients with progressive [19, 22, 27] or refractory disease [23]. Thus, it is likely that none of these mutations are relevant toward determining an indolent disease course.
In addition, in our US-CLL no clonal mutation of TP53, NOTCH1, SF3B1 and BIRC3 genes [14–16, 27, 29] was found, including the US-CLL collected for the gene expression study (supplementary Table S4, available at Annals of Oncology online). Moreover, none of the discovery cohort cases showed ATM mutations or any established/putative driver mutation responsible of disease progression in mutated IGHV CLL [27].
Unexpectedly, we found 2 US-CLL cases harboring TP53 mutations with a low VAF. These cases mimic the rare clonal TP53 disrupted patients with a long-term stable disease, allowed by the absence of other poor risk prognostic factors and by the absence of treatment selective pressure [6, 26, 30–33].
CNA analysis of US-CLL documented del(13q) only in 31%, no CNA in 38% and a load of 1 lesion/case, less than what reported in the largest studies based on SNP 6.0 arrays, with very few CNAs in common [7, 8, 20]. No other recurrent lesion was detected.
Overall, the low load of mutations/CNAs and the absence of unfavorable mutations/CNAs—even in long follow-up samples—could reflect the genomic stability of US-CLL cases that, over the years, do not tend to accumulate genetic lesions driving disease progression, thus maintaining a long-lasting indolent disease.
We also investigated the transcriptional profile of US-CLL compared with that of non-US-CLL who experienced a disease progression in spite of good conventional prognostic factors. The expression levels of eight genes, quantified on an extended cohort of cases using an innovative platform (ddPCR), allowed to build a six gene-based decision-tree capable of recognizing among CLL patients with a favorable immunogenetic profile those with an US clinical course, with an accuracy of 95%. This predictive model, potentially capable of identifying US patients at diagnosis, deserves further validation on an independent cohort of cases.
In the current CLL guidelines, ‘watch and wait’ still remains the standard of care for early stage and asymptomatic CLL patients irrespective of prognostic factors [13]. Nevertheless, early treatment intervention is a tempting investigational scenario in the era of novel drugs. A number of proposed prognostic algorithms—i.e. smouldering CLL, CLL-IPI, MDACC 2007, MDACC 2011, Barcelona-Brno, O-CLL1 score—can identify low-risk CLL patients; our US-CLL patients meet the criteria of low-risk whatever algorithm is applied [34, 35]. However, even within low-risk CLL there is still a sizable proportion of patients experiencing ‘early’ progression. This had been recently highlighted by Molica et al. [35] who compared six scoring systems—CLL-IPI, MDACC 2007, MDACC 2011, GCLLSG, Barcelona-Brno, O-CLL1—and found that 22%–35% of prediction by these algorithms fails when applied to CLL in early phase. Our six-gene expression algorithm could fill this gap, complementing genetics in identifying truly low-risk CLL patients. Beside other relevant clinical implications including patients’ counseling at diagnosis and subsequent monitoring, it could further refine the current prognostic algorithms allowing to identify within low-risk CLL patients those who will not progress for decades and might never require treatment and those who may benefit from experimental early intervention.
Funding
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) Special Program Molecular Clinical Oncology, 5x1000, No. 10007, Milan, Italy; Progetto Giovani Ricercatori 2010: Policlinico di Modena (GR-2010-2313609); Ricerca Finalizzata (RF-2011-02349712), Ministry of Health, Rome, Italy to GMR, AC, RF and GG; Progetti di Rilevante Interesse Nazionale (PRIN) (2015ZMRFEA) to RF, GG and AC; Gilead Fellowship Program 2016 to AG; Precision Medicine Fellowship (UL1 TR000040) to JW.
Disclosure
The authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Fabbri G, Dalla-Favera R.. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer 2016; 16(3): 145–162 (Review). [DOI] [PubMed] [Google Scholar]
- 2. Pospisilova S, Gonzalez D, Malcikova J. et al. ; European Research Initiative on CLL (ERIC). ERIC recommendations on TP53 mutation analysis in chronic lymphocytic leukemia. Leukemia 2012; 26(7): 1458–1461. [DOI] [PubMed] [Google Scholar]
- 3. Guarini A, Marinelli M, Tavolaro S. et al. ATM gene alterations in chronic lymphocytic leukemia patients induce a distinct gene expression profile and predict disease progression. Haematologica 2012; 97(1): 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Foà R, Del Giudice I, Guarini A. et al. Clinical implications of the molecular genetics of chronic lymphocytic leukemia. Haematologica 2013; 98(5): 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rossi D, Rasi S, Spina V. et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood 2013; 121(8): 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rossi D, Khiabanian H, Spina V. et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood 2014; 123(14): 2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ouillette P, Collins R, Shakhan S. et al. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood 2011; 118(11): 3051–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edelmann J, Holzmann K, Miller F. et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood 2012; 120(24): 4783–4794. [DOI] [PubMed] [Google Scholar]
- 9. Guarini A, Gaidano G, Mauro FR. et al. Chronic lymphocytic leukemia patients with highly stable and indolent disease show distinctive phenotypic and genotypic features. Blood 2003; 102(3): 1035–1041. [DOI] [PubMed] [Google Scholar]
- 10. Capello D, Guarini A, Berra E. et al. Evidence of biased immunoglobulin variable gene usage in highly stable B-cell chronic lymphocytic leukemia. Leukemia 2004; 18(12): 1941–1947. [DOI] [PubMed] [Google Scholar]
- 11. Del Giudice I, Mauro FR, De Propris MS. et al. White blood cell count at diagnosis and immunoglobulin variable region gene mutations are independent predictors of treatment-free survival in young patients with stage A chronic lymphocytic leukemia. Haematologica 2011; 96(4): 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Agathangelidis A, Darzentas N, Hadzidimitriou A. et al. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood 2012; 119(19): 4467–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eichhorst B, Robak T, Montserrat E, ESMO Guidelines Committee et al. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2015; 26(Suppl 5): v78–v84. [DOI] [PubMed] [Google Scholar]
- 14. Rossi D, Bruscaggin A, Spina V. et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood 2011; 118(26): 6904–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rossi D, Rasi S, Fabbri G. et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood 2012; 119(2): 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rossi D, Fangazio M, Rasi S. et al. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood 2012; 1, 2854–2862. [DOI] [PubMed] [Google Scholar]
- 17. Messina M, Chiaretti S, Tavolaro S. et al. Protein kinase gene expression profiling and in vitro functional experiments identify novel potential therapeutic targets in adult acute lymphoblastic leukemia. Cancer 2010; 116(14): 3426–3437. [DOI] [PubMed] [Google Scholar]
- 18. Del Giudice I, Chiaretti S, Tavolaro S. et al. Spontaneous regression of chronic lymphocytic leukemia: clinical and biologic features of 9 cases. Blood 2009; 114(3): 638–646. [DOI] [PubMed] [Google Scholar]
- 19. Puente XS, Pinyol M, Quesada V. et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; 475(7354): 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang L, Lawrence MS, Wan Y. et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 2011; 365(26): 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quesada V, Conde L, Villamor N. et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 2012; 44(1): 47–52. [DOI] [PubMed] [Google Scholar]
- 22. Landau DA, Tausch E, Taylor-Weiner AN. et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015; 526(7574): 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Messina M, Del Giudice I, Khiabanian H. et al. Genetic lesions associated with chronic lymphocytic leukemia chemo-refractoriness. Blood 2014; 123(15): 2378–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morin RD, Assouline S, Alcaide M. et al. Genetic landscapes of relapsed and refractory diffuse large B-cell lymphomas. Clin Cancer Res 2016; 22(9): 2290–2300. [DOI] [PubMed] [Google Scholar]
- 25. Kataoka K, Nagata Y, Kitanaka A. et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet 2015; 47(11): 1304–1315. [DOI] [PubMed] [Google Scholar]
- 26. Landau DA, Carter SL, Stojanov P. et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013; 152(4): 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rose-Zerilli MJ, Gibson J, Wang J. et al. Longitudinal copy number, whole exome and targeted deep sequencing of ‘good risk’ IGHV-mutated CLL patients with progressive disease. Leukemia 2016; 30(6): 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Puente XS, Beà S, Valdés-Mas R. et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015; 526(7574): 519–524. [DOI] [PubMed] [Google Scholar]
- 29. Baliakas P, Hadzidimitriou A, Sutton LA. et al. ; European Research Initiative on CLL (ERIC). Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia 2015; 29(2): 329–336. [DOI] [PubMed] [Google Scholar]
- 30. Malcikova J, Stano-Kozubik K, Tichy B. et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia 2015; 29(4): 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Best OG, Gardiner AC, Davis ZA. et al. A subset of Binet stage A CLL patients with TP53 abnormalities and mutated IGHV genes have stable disease. Leukemia 2009; 23(1): 212–214. [DOI] [PubMed] [Google Scholar]
- 32. Delgado J, Salaverria I, Baumann T. et al. Genomic complexity and IGHV mutational status are key predictors of outcome of chronic lymphocytic leukemia patients with TP53 disruption. Haematologica 2014; 99(11): e231–e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu L, Kim HT, Kasar SN. et al. Survival of Del17p CLL depends on genomic complexity and somatic mutation. Clin Cancer Res 2017; 23(3): 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. International CLL-IPI working group. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol 2016; 17(6): 779–790. [DOI] [PubMed] [Google Scholar]
- 35. Molica S, Giannarelli D, Mirabelli R. et al. Reliability of six prognostic models to predict time-to-first-treatment in patients with chronic lymphocytic leukaemia in early phase. Am J Hematol 2017; 92(6): E91–E93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.