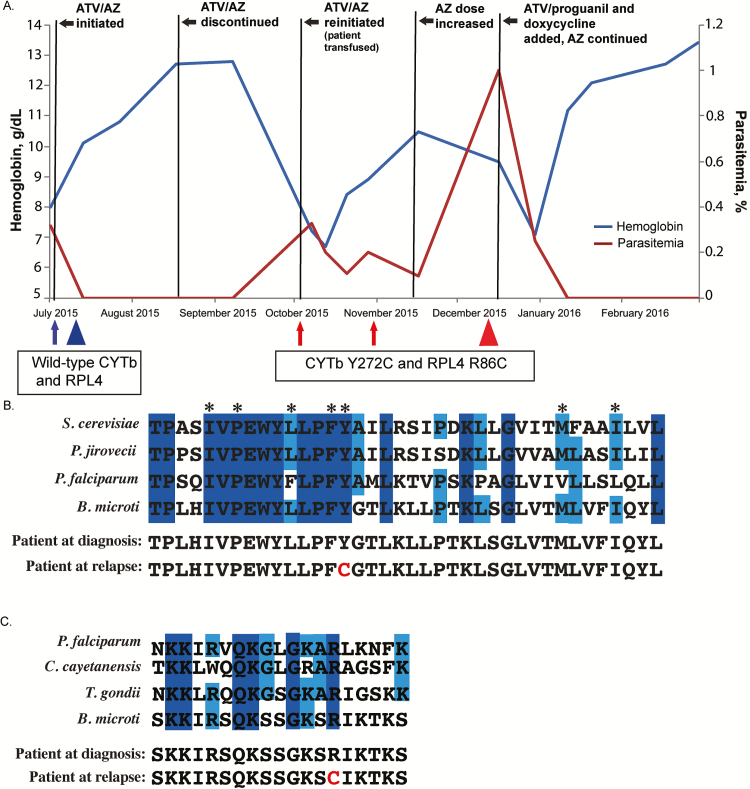
Figure 1.
A, Case patient’s clinical and treatment course, including hemoglobin values and parasitemia. Five residual samples were collected at different times during the course of infection. Arrows and triangles indicate when samples were obtained; arrows denote samples from which only cytb was sequenced; triangles, samples from which cytb and rpl4 were sequenced. Blue represents the presence of wild-type cytb and rpl4 alleles at initial diagnosis; red, the presumed resistant alleles after relapse following a 6-week course of atovaquone/azithromycin (ATV/AZ). B, Amino acid residue sequence conservation of the ATV-binding pocket. Alignment of Babesia microti and model organisms’ cytochrome b (CYTb) protein sequences: Saccharomyces cerevisiae, Pneumocystis jirovecii, and Plasmodium falciparum. Residues comprise the QO site. Dark- and light-blue shading indicate residues that are 100% or 75% conserved, respectively. Asterisks indicate residues previously described as harboring important side-chain interactions with ATV. Amino acid alignment of the case patient’s predicted CYTb sequences before and after relapse are shown, with the amino acid change at position 272 noted in red . C, Amino acid alignment of putative AZ-binding pocket of ribosomal protein L4 (RPL4), comparing B. microti and model organisms, P. falciparum, Cyclospora cayetanensis, and Toxoplasma gondii. Alignment demonstrates the patient’s predicted RPL4 sequence before and after treatment. Dark- and light-blue shading indicate residues that are 100% or 75% conserved, respectively. The amino acid change corresponding to presumed resistance is highlighted in red . All sequences were obtained from the EuPathDB database (eupathdb.org).