We identified single-nucleotide polymorphisms (SNPs) of stemness-related genes, including CD44 (rs9666607), ABCC1 (rs35605 and rs212091) and GDF15 (rs1058587) that were associated with prostate cancer survival and were predicted to regulate RNA splicing, microRNA and oncogenic signaling.
Abstract
Prostate cancer (PCa) is a clinically and molecularly heterogeneous disease, with variation in outcomes only partially predicted by grade and stage. Additional tools to distinguish indolent from aggressive disease are needed. Phenotypic characteristics of stemness correlate with poor cancer prognosis. Given this correlation, we identified single-nucleotide polymorphisms (SNPs) of stemness-related genes and examined their associations with PCa survival. SNPs within stemness-related genes were analyzed for association with overall survival of PCa in the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial. Significant SNPs predicted to be functional were selected for linkage disequilibrium analysis and combined and stratified analyses. Identified SNPs were evaluated for association with gene expression. SNPs of CD44 (rs9666607), ABCC1 (rs35605 and rs212091) and GDF15 (rs1058587) were associated with PCa survival and predicted to be functional. A role for rs9666607 of CD44 and rs35605 of ABCC1 in RNA splicing regulation, rs212091 of ABCC1 in miRNA binding site activity and rs1058587 of GDF15 in causing an amino acid change was predicted. These SNPs represent potential novel prognostic markers for overall survival of PCa and support a contribution of the stemness pathway to PCa patient outcome.
Introduction
Prostate cancer (PCa) is the most common cancer and second leading cause of cancer-related death in men living in the USA (1). The natural course of PCa exhibits tremendous clinical heterogeneity; therefore, prognostic tools distinguishing indolent from aggressive disease are urgently needed to delineate patients at greater risk of progressing on active surveillance or through localized therapy. In addition, these tools have the potential to increase our understanding of the molecular mechanisms underlying aggressive disease and to rationalize specific targeting of particular oncogenic signaling pathways in selected patients. Such tools to distinguish and target aggressive disease may also impact PCa disparities and the more aggressive characteristics of PCa in African American men (2). Current prognostic tools for PCa use non-specific host (age, race, comorbidities), tumor [prostate-specific antigen (PSA), Gleason score] and post-treatment characteristics (morphological and molecular markers) (3). Pre- and post-operative nomograms employ combinations of the aforementioned characteristics with additional molecular markers (4).
Much evidence suggests that cells having a stemness phenotype play important roles in cancer initiation, progression and lack of treatment efficacy, and thus cancer-related death (5). Cells within the prostate having a stemness phenotype, characterized by an ability to proliferate, self-renew and give rise to the cellular heterogeneity of the tissue, have been identified (6–9). In addition to the benign cells within the prostate that have a stemness phenotype, PCa cells possessing this phenotype have also been identified (10,11). As is generally a characteristic of cancer cells having a stemness phenotype, PCa cells having this phenotype tend to metastasize and be resistant to therapeutic agents, including androgen deprivation therapy, chemotherapy and radiotherapy (12–14). Consistent with this phenotype, the gene expression signature of these cells is predictive of poor prognosis. Gene expression profiling studies have revealed genes upregulated in PCa progenitor cells, including epidermal growth factor receptor, hedgehog, Wnt/β-catenin, Notch, hyaluronan/CD44, stromal cell-derived factor-1/chemokine receptor 4, genes playing a role in glycolytic metabolism, Akt, NF-κB, hypoxia-inducible factors and embryonic stem cell-like transcription factors that play a role in stemness, including Oct3/4, Sox2, Nanog and Bmi-1 (15). Despite the presence and potential of prostate cells having a stemness phenotype to portend a poor prognosis, and the utmost importance of such a phenotype in populations at the greatest risk for developing aggressive disease, none of the current prognostic tools for PCa directly reflect stemness.
It is well established that germline single-nucleotide polymorphisms (SNPs) are associated with cancer risk, confirming the significance of genetic variation as a molecular mechanism underlying carcinogenesis. PCa is no exception to this paradigm, and a meta-analysis using high-density SNP genotyping data from nine studies, including subjects of European, African, Japanese and Latino ancestry, has identified 23 SNPs associated with PCa risk (16). In addition, SNPs associated with early onset of PCa and with aggressive PCa have also been identified (17,18). In the post-genomic and post-GWAS era, it is possible to take a hypothesis-driven, targeted pathway-based, multigene approach to identify genetic variation in an oncogenic signaling pathway and its association with cancer risk. Recently, we have identified SNPs in stemness-related genes that were significantly associated with racial disparities in susceptibility to PCa and were predicted to function in regulation of RNA splicing (19). Far fewer studies have revealed germline SNPs associated with PCa prognosis, in part because of the challenge of obtaining sufficient sample sizes with clinical annotation. Two genome-wide association studies (GWASs) have demonstrated the potential of germline SNPs to have an effect on PCa prognosis, as these studies have used the ProtecT data and the Korean Cancer Prevention Study-II and Korean Genome Epidemiology Study to identify SNPs associated with the PSA level (20,21). A limited number of smaller studies have also demonstrated the potential for germline SNPs to predict PCa prognosis; therefore, it is important to identify SNPs associated with early biochemical recurrence, PSA levels, tumor and prostate volume and PCa aggressiveness/Gleason score (22–26).
Given the previous identification of germline SNPs that are associated with PCa prognosis, it is likely that identifying genetic variation in additional oncogenic signaling pathways will lead to novel tools for PCa prognosis. In the post-GWAS era, it is possible to take a hypothesis-driven, targeted pathway-based, multigene approach to identify genetic variation in an oncogenic signaling pathway and its association with cancer survival. Given the potential of prostate cells having a stemness phenotype to portend a poor prognosis, it is likely that genetic variation contributing to this phenotype could also serve as novel precision biomarkers that have prognostic significance for PCa in distinguishing aggressive from indolent disease at the time of screening and diagnosis and/or as novel molecular targets for developmental therapeutics against aggressive PCa (19). To elucidate such genetic variation, we applied a hypothesis-driven, targeted pathway-based, multigene approach to identify SNPs of stemness-related genes and examine their associations with survival of PCa patients using available GWAS genotyping data from the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO) (27). Compared with GWAS analyses, the approach used here has the advantages of including biologically relevant targets a priori, allowing fewer SNPs to be honed in on for analyses and enabling expression quantitative trait loci (eQTL) analyses to assess the functional relevance of SNPs under investigation. It should be noted that the typical highly stringent GWAS significance threshold is not applicable to this targeted pathway-based approach because the number of SNPs tested is much lower.
Materials and methods
Study population
The present study included 1150 PCa cases that were diagnosed among men enrolled in the PLCO cohort (27). The PLCO originally enrolled 77500 men and 77500 women aged 55–74 years. It is a National Cancer Institute (NCI)-funded, multicenter, randomized trial focused on screening for cancer at 10 medical centers in the USA between 1993 and 2011. The PLCO collected blood specimens from the first screening visit, gathered extensive information about each participant and followed all participants for at least 13 years after enrollment. Genomic DNA extracted from the blood samples was genotyped using Illumina HumanHap300v1.1 and HumanHap250Sv1.0 (dbGaP accession: phs000207.v1.p1) (28). The present study includes genotype data from 1150 non-Hispanic white PCa patients. Tumor staging was determined according to the fifth edition American Joint Committee on Cancer (AJCC) staging system. The follow-up time was defined from PCa diagnosis to the date of last follow-up or time of death. We used overall survival (OS) of PCa as the primary end point of the present study. The institutional review boards of each participating institution approved the PLCO and the use of biospecimens for further research, and all subjects signed a written informed consent. The current analyses were conducted after application to and approval from the NIH/NCI.
Gene and SNP selection
Based on the online database Genecards (http://www.genecards.org//), 25 stemness-related genes that reportedly play a role in PCa were selected, using the search terms ‘prostate cancer stem cell’ (Supplementary Table S1, available at Carcinogenesis Online). Genotyped SNPs within these genes and their ±2 kb flanking regions were selected for association analysis. There were available data for 635 genotyped SNPs of the 25 genes in PLCO. SNPs in the GWAS data set were further selected by using the following criteria: SNPs located on autosomal chromosomes; minor allelic frequency ≥ 5%; a genotyping rate ≥ 95% and Hardy–Weinberg equilibrium ≥ 1 × 10−6. As a result, 635 typed SNPs of the 25 genes were extracted from the PLCO PCa GWAS data (dbGaP accession: phs000207.v1.p1). Nineteen typed SNPs of 10 of the 25 genes showed an association with PCa OS and passed multiple testing correction by the false-positive report probability (FPRP) method that is independent of the number of multiple tests. These 10 gene regions were further imputed, filtering the imputed SNPs with the criteria of minor allelic frequency ≥ 5%, genotyping rate ≥ 95% and Hardy–Weinberg equilibrium ≥ 1 × 10−6 by using sequencing data from the 1000 Genomes Project to identify untyped functional SNPs that are linked to the significant tagSNPs. The functional relevance of these SNPs was predicted by SNPinfo and RegulomeDB, which are publically available online tools. SNPinfo incorporates functional predictions of protein structure, gene regulation, RNA splicing and microRNA binding (29). RegulomeDB was used to identify the SNPs with previously reported links to eQTL labeled within a score of ‘1’ (30).
Statistical analysis
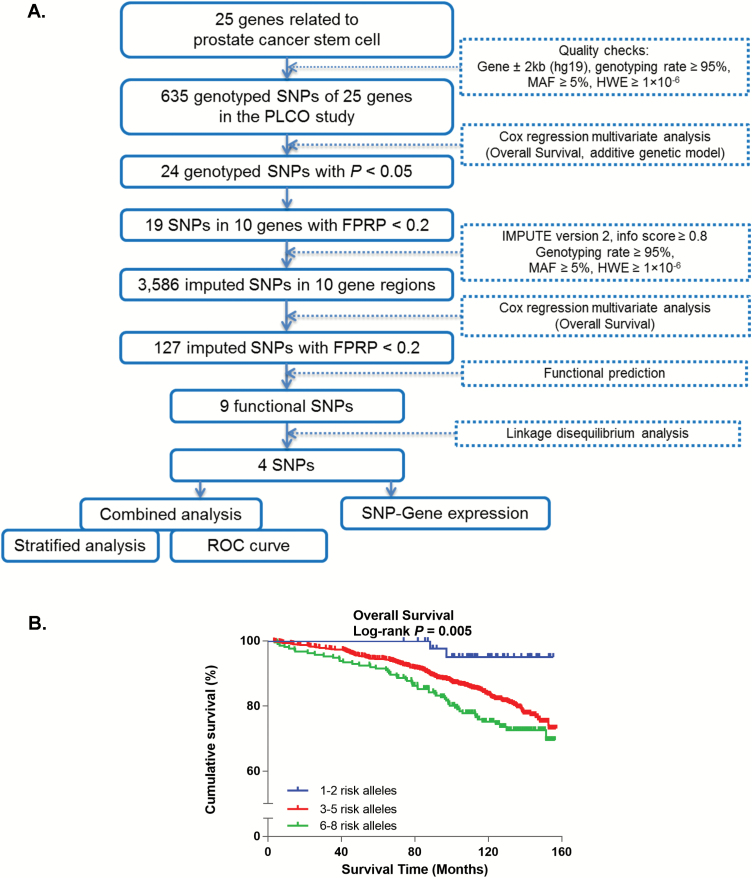
Figure 1A provides a flowchart that outlines the analyses. Briefly, Cox proportional hazards regression models were completed for each of the selected SNPs of the 25 PCa stem cell-related genes. For multiple testing correction, the FPRP approach was used with a cutoff value of 0.2 to lower the probability of false-positive findings, which depends on the observed P value, the statistical power and the prior probability, rather than the number of tests (31). This is because many of the SNPs selected in the GWAS data set are in linkage disequilibrium (LD), not independent per se, particularly for those obtained from imputation. Based on the typed SNPs on 500 kb flanking regions of the selected gene regions from the GWAS data set, genotype imputation was performed with IMPUTE2 using multi-population reference panels from the 1000 Genomes Project Phase 3 (2014 release) (32). Imputed SNPs with info value ≥ 0.8 were qualified for further analysis. Further filtering was completed using functional prediction utilizing SNPinfo and eQTL annotation of RegulomeDB. Pairwise LD was estimated by using the data from Europeans in the 1000 Genomes Project Phase 3. The number of effect genotypes was summarized to evaluate the combined effects of all the independent and significant SNPs.
Figure 1.
(A) Overall flowchart. (B) Kaplan–Meier survival curves of prostate cancer patients, according to combined risk alleles of the four independent and significant SNPs in low-, median-, and high-risk groups. MAF, minor allele frequency; HWE, Hardy–Weinberg equilibrium.
Kaplan–Meier curves and log-rank tests were used to evaluate the associations between genotypes and OS. The heterogeneity of associations between subgroups in stratified analyses was assessed using the χ2-based Q-test. Cox regression models were used to estimate the hazard ratio (HR) and 95% confidence interval (CI) for the associations of demographic and clinical characteristics with OS. Associations between SNPs and OS (in additive genetic models) were obtained by both univariate and multivariable Cox regression analyses performed by using the GenABEL package of R software, with adjustments for age, Gleason score, stage and primary treatment (33). In selecting our final multivariable model, we combined risk alleles of the final set of independent and significant SNPs into genetic ‘scores’ to test their joint effects on PCa survival. All patients were allocated into eight groups, with one to eight risk alleles. Final model selection was completed based on the lowest Akaike information criterion (34). A time-dependent receiver-operating characteristic (ROC) analysis was performed to calculate area under curve (AUC) of SNPs and clinical characteristics by using ‘survAUC’ package of R software (35). As an internal validation, the final multivariate models were replicated in 1000 bootstrap samples, where in each cohort sampled, a random two-third training set and one-third testing set were analyzed with an AUC computed and then averaged over the 1000 samples. We report the mean bootstrap AUC and 95% bootstrap CIs of the AUC for 3-, 5- and 10-year survival, as well as the bootstrap P value, comparing the model with clinical variables alone with clinical plus genetic variables in 1000 bootstrap training sets.
Correlations between SNPs and mRNA expression of the corresponding genes were analyzed by using general linear regression with an additive genetic model in R software. Genotype data and normalized mRNA expression levels were obtained from lymphoblastoid cell lines from 716 unrelated individuals of the HapMap 3 Project using Illumina Human-6 v2 Expression BeadChip, including 107 Northern Europeans from Utah (CEU), 242 Asians (CHB), 41 individuals of Mexican (MXL) ancestry and 326 individuals of African (YRI) ancestry (36). The raw expression data were normalized on a log2 scale using a quantile method, as described previously in the published article from the HapMap 3 Project. All statistical analyses were calculated using SAS software (version 9.1.3; SAS Institute, Cary, NC) unless otherwise specified.
Results
Basic characteristics of the study population
The overall workflow of the present study is summarized in Figure 1A. Basic characteristics of the 1150 non-Hispanic white PCa patients from the PLCO are described in Table 1. The median age of the patients was 67 years. Of these 1150 patients, 215 (18.7%) were deceased at the last follow-up (Table 1). The median follow-up time was 121.7 months. In multivariate analyses, five of the six selected variables were found to be significantly associated with PCa OS. These variables were age at diagnosis (HR = 1.80, >67 versus ≤67), Gleason score (HR = 2.52, ≥8 versus 2–6), tumor stage (HR = 1.76, III/IV versus I/II), aggressiveness (HR = 1.89, non-aggressive versus aggressive) and treatment (HR = 2.06, 1.56, 4.28 and 2.68 for radiotherapy alone, radiotherapy + hormone therapy, hormone therapy alone and other treatments versus radical prostatectomy, respectively).
Table 1.
Clinical characteristics of the prostate cancer PLCO study in overall survival
| Characteristics | Frequency | Univariate analysis | Multivariate analysisa | |||
|---|---|---|---|---|---|---|
| All | Deaths (%) | HR (95% CI) | P | HR (95% CI) | P | |
| Overall | 1150 | 215 (18.7) | ||||
| Age (years) | ||||||
| Median (range) | 67 (55–81) | |||||
| <67 | 544 | 70 (12.9) | 1.00 | 1.00 | ||
| ≥67 | 606 | 145 (23.9) | 2.24 (1.68–2.98) | <0.001 | 1.80 (1.34–2.42) | <0.001 |
| PSA before diagnosis (ng/ml) | ||||||
| Median (range) | 6.1 (0.05–1137) | |||||
| <6.1 | 572 | 89 (15.6) | 1.00 | 1.00 | ||
| ≥6.1 | 578 | 126 (21.8) | 1.38 (1.05–1.81) | 0.021 | 0.98 (0.74–1.30) | 0.887 |
| Gleason score | ||||||
| 2 ≥ & ≤ 6 | 567 | 96 (16.9) | 1.00 | 1.00 | ||
| =7 | 464 | 76 (16.4) | 1.19 (0.88–1.61) | 0.254 | 1.23 (0.90–1.70) | 0.199 |
| ≥8 | 114 | 41 (36.0) | 3.13 (2.17–4.53) | < 0.001 | 2.52 (1.68–3.77) | <0.001 |
| Missing | 5 | |||||
| Stage | ||||||
| I/II | 913 | 153 (16.7) | 1.00 | 1.00 | ||
| III/IV | 237 | 62 (26.2) | 1.62 (1.20–2.17) | 0.002 | 1.76 (1.26–2.46) | 0.001 |
| Aggressivenessb | ||||||
| Non-aggressive | 489 | 78 (16.0) | 1.00 | 1.00 | ||
| Aggressive | 659 | 137 (20.8) | 1.65 (1.24–2.18) | 0.001 | 1.89 (1.40–2.56) | <0.001 |
| Missing | 2 | |||||
| Types of treatments | ||||||
| Radical prostatectomy | 614 | 75 (12.2) | 1.00 | 1.00 | ||
| Radiotherapy alone | 194 | 46 (23.7) | 2.00 (1.39–2.89) | <0.001 | 2.06 (1.39–3.06) | <0.001 |
| Radiotherapy + endocrine therapy | 202 | 40 (19.8) | 1.96 (1.34–2.88) | 0.001 | 1.56 (1.03–2.36) | 0.035 |
| Endocrine therapy alone | 54 | 29 (53.7) | 7.21 (4.68–11.09) | <0.001 | 4.28 (2.64–6.92) | <0.001 |
| Other treatments | 86 | 25 (29.1) | 2.53 (1.61–3.98) | <0.001 | 2.68 (1.65–4.36) | <0.001 |
aMultivariate Cox regression analyses were adjusted for age, Gleason score, PSA level, stage and primary treatments. In subgroup of ‘Aggressiveness’, we included age, PSA level and primary treatment for adjustments.
bNon-aggressive: cases with a Gleason score < 7 and stage < III; Aggressive: cases with a Gleason score ≥ 7 or stage ≥ III.
Multivariate analyses of associations between SNPs and PCa OS
Multivariate Cox models were used in the single locus analysis to assess associations of 635 genotyped SNPs with OS in the presence of age, Gleason score, stage and primary treatment (as summarized in the Manhattan plot, Supplementary Figure S1, available at Carcinogenesis Online). Of these 635 SNPs, 24 SNPs were individually significantly associated with OS at P < 0.05 in an additive genetic model. After applying the FPRP for noteworthy SNPs, 19 SNPs in 10 genes (TP63, ITGA1, EGFR, MET, ALDH1A1, ITGB1, CD44, ABCC1, GDF15 and ERG) remained statistically significant, with an FPRP < 0.2 (Supplementary Table S2, available at Carcinogenesis Online). Then, we performed the imputation for potentially functional SNPs in the 10 genes (Supplementary Table S3, available at Carcinogenesis Online). Of the 3484 imputed SNPs, 9 SNPs with potential functions as predicted by SNPinfo and eQTL annotation of RegulomeDB were finally identified among 127 SNPs that remained significantly associated with PCa OS in the single locus analysis after applying an FPRP < 0.2 for noteworthy SNPs (Supplementary Table S4, available at Carcinogenesis Online).
LD analysis of the nine predicted functional SNPs
In the LD analysis of the nine imputed SNPs predicted to be functional, five SNPs of ABCC1 (rs35604, rs35605, rs35607, rs35610 and rs35613) and two SNPs of GDF15 (rs1058587 and rs16982345) were in high LD, respectively (all r2 > 0.8) (Supplementary Figure S2, available at Carcinogenesis Online). The five SNPs of ABCC1 mentioned above were all in low LD with rs212091 (all r2 < 0.2) located in 3′UTR. Compared with the high LD SNPs of the corresponding genes, rs35605 of ABCC1 and rs1058587 of GDF15 exhibited more functional relevance, and both were located in exonic regions (Table 2). Therefore, rs35605, rs1058587 and two other SNPs (rs212091 and rs9666607) were chosen as independent SNPs for additional analyses. Regional association plots of the regions around the four SNPs are shown in Supplementary Figure S3, available at Carcinogenesis Online.
Table 2.
The four independent and significant SNPs with potential functions
| SNP | Gene | Chr. | Position (hg19) | Location | Allelea | EAF | SNPinfob | RegulomeDBc | HR (95% CI)d | P d | FPRPd |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs9666607 | CD44 | 11 | 35226155 | Exon | G/A | 0.31 | Splicing | 5 | 1.28 (1.04–1.58) | 0.018 | 0.141 |
| rs35605 | ABCC1 | 16 | 16162019 | Exon | C/T | 0.16 | Splicing | 1f | 0.71 (0.53–0.94) | 0.018 | 0.138 |
| rs212091 | ABCC1 | 16 | 16236650 | 3′UTR | T/C | 0.15 | miRNA | 7 | 0.58 (0.43–0.80) | 0.001 | 0.009 |
| rs1058587 | GDF15 | 19 | 18499422 | Exon | C/G | 0.26 | nsSNP | 4 | 1.29 (1.05–1.59) | 0.015 | 0.117 |
Chr., chromosome; EAF, effect allele frequency; FPRP, false positive report probability; nsSNP, non-synonymous SNP; UTR, untranslated region.
aReference/effect allele.
cRegulomeDB, http://regulome.stanford.edu/. SNPs with predicted scores of ‘1’ were considered as functional.
dMultivariate Cox regression analyses of an additive genetic model were adjusted for age, Gleason score, stage and primary treatments.
Combined and stratified analyses of the four independent SNPs
The minor alleles of rs35605 and rs212091 of ABCC1 were found to be associated with better OS of PCa, with a variant-allele attributed HR of 0.71 (95% CI = 0.53–0.94, P = 0.018) and 0.58 (95% CI = 0.43–0.80, P = 0.001), respectively (Table 2). Compared with their corresponding reference genotypes in a dominant genetic model, patients with TC and TT genotypes of rs35605 and TC and CC genotypes of rs212091 had a decreased risk of death (HR = 0.71, 95% CI = 0.51–0.97 and P = 0.034; HR = 0.60, 95% CI = 0.43–0.84 and P = 0.003, respectively; Supplementary Table S5, available at Carcinogenesis Online). Meanwhile, the minor alleles of rs9666607 of CD44 and rs1058587 of GDF15 were associated with a worse OS from PCa in an additive genetic model, with an HR of 1.28 (95% CI = 1.04–1.58, P = 0.018) and 1.29 (95% CI = 1.05–1.59, P = 0.015), respectively (Table 2). Compared with the reference genotypes in a recessive genetic model, patients with risk genotypes of two SNPs had an increased risk of death (HR = 1.86 and P = 0.002 for rs9666607; HR = 1.76 and P = 0.015 for rs1058587, respectively; Supplementary Table S5, available at Carcinogenesis Online). In addition, we determined the associations between the four SNPs and PCa-specific survival. Although no trend reached statistical significance, CD44 rs9666607, ABCC1 rs35605 and ABCC1 rs212091 showed the same trend effecting disease-specific survival that was shown effecting OS (Supplementary Table S6, available at Carcinogenesis Online).
We assessed five models that combined the risk alleles utilizing different groupings (Table 3 and Supplementary Table S7, available at Carcinogenesis Online). Based on the Akaike information criterion, the model that used four groups of all patients having 1–2, 3, 4–5 and 6–8 risk alleles, respectively, was preferred. In this model, an increase in per-unit risk score was significantly associated with survival after adjustments (P = 5.30 × 10−7, Table 3). More specifically, compared with the lowest risk group, the other three risk groups of 3, 4–5 and 6–8 risk alleles had an HR of 4.81, 8.35 and 11.80-fold increased risk of death, respectively, with 95% CI and P value of 1.10–21.00 and 0.037; 2.04–34.19 and 0.003; and 2.84–49.04 and 0.001, respectively (Table 3). To visualize the HR effects, we present Kaplan–Meier survival curves of the association between OS and genotypes on the four SNPs in Supplementary Figure S4, available at Carcinogenesis Online, and the combined effects in Figure 1B and Supplementary Figure S5, available at Carcinogenesis Online.
Table 3.
Combined analysis of the four independent and significant SNPs in four risk groups
| Number of risk allelesa | Frequency | Multivariate analysisb | AICc | ||
|---|---|---|---|---|---|
| All | Deaths (%) | HR (95% CI) | P | ||
| 1–2 | 46 | 2 (4.3)d | 1.00 | ||
| 3 | 158 | 19 (12.0) | 4.81 (1.10–21.00) | 0.037 | |
| 4–5 | 709 | 138 (19.5) | 8.35 (2.04–34.19) | 0.003 | |
| 6–8 | 213 | 53 (24.9) | 11.80 (2.84–49.04) | 0.001 | |
| Trend | 5.30E−07 | 2666.68 | |||
AIC, Akaike information criterion.
aRisk alleles were rs9666607 A, rs35605 C, rs212091 T and rs1058587 G.
bMultivariate Cox regression analyses were adjusted for age, Gleason score, stage and primary treatments.
cAIC in the trend model of multivariate Cox regression analyses.
dTwo patients reached the end point were both with two risk alleles.
In subgroup analyses of patients with different risk scores, which were stratified by age, PSA before diagnosis, Gleason score, stage, aggressiveness or types of primary treatments, we found no significant evidence for heterogeneity across strata (all P for heterogeneity > 0.05, Supplementary Table S8, available at Carcinogenesis Online). For each independent SNP in stratified analysis, we observed heterogeneity in the age group for ABCC1 rs212091 (P for heterogeneity = 0.017, Supplementary Table S9, available at Carcinogenesis Online). In the subgroup of older age, patients carried the protective allele C of rs212091 showed a better survival, which means the allele T was associated with an increased risk of death (Supplementary Table S9, available at Carcinogenesis Online).
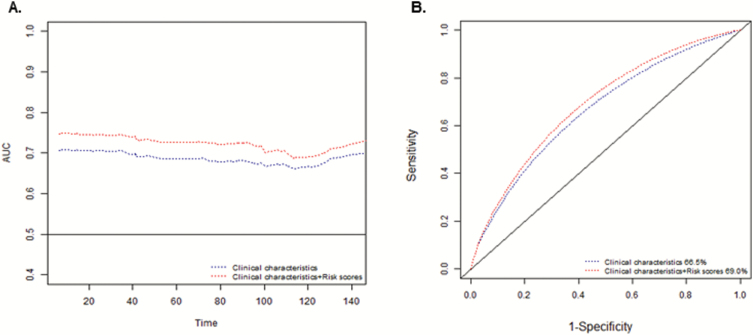
ROC curve and internal validation
We further evaluated combined risk scores for their potential to predict PCa OS by a time-dependent ROC. As shown in Figure 2A, the AUC based on both trichotomized risk scores and clinical characteristics was greater than that with only clinical characteristics at different time points. The AUC of the 10-year survival models increased from 66.5% to 69.0% after adding the genetic scores to the clinical characteristics (Figure 2B). To obtain a more accurate estimate of the predictive performance, we also applied the internal validation method (bootstrap) to estimate the AUC and 95% CI. The bootstrap mean AUC and 95% CIs for 3-year survival were 73.3 (68.5–78.2) for clinical variables only and 75.5 (70.4–80.5) for clinical plus genetic scores, bootstrap P value = 0.032; similarly, for 5-year survival, 72.2 (68.0–76.4) for clinical variables only and 74.2 (69.8–78.4) for clinical plus genetic, bootstrap P value = 0.01; and finally, for 10-year survival, 69.7 (65.9–73.5) for clinical variables only and 71.7 (68.1–75.2) for clinical plus genetic scores, bootstrap P value = 0.002.
Figure 2.
ROC curves for prediction of prostate cancer survival based on only clinical characteristics (age, Gleason score and tumor stage) or additionally combined effect genotypes, (A) time-dependent AUC and (B) ROC curves corresponding to the time point of 10 years.
The four independent SNPs and mRNA expression of the related genes
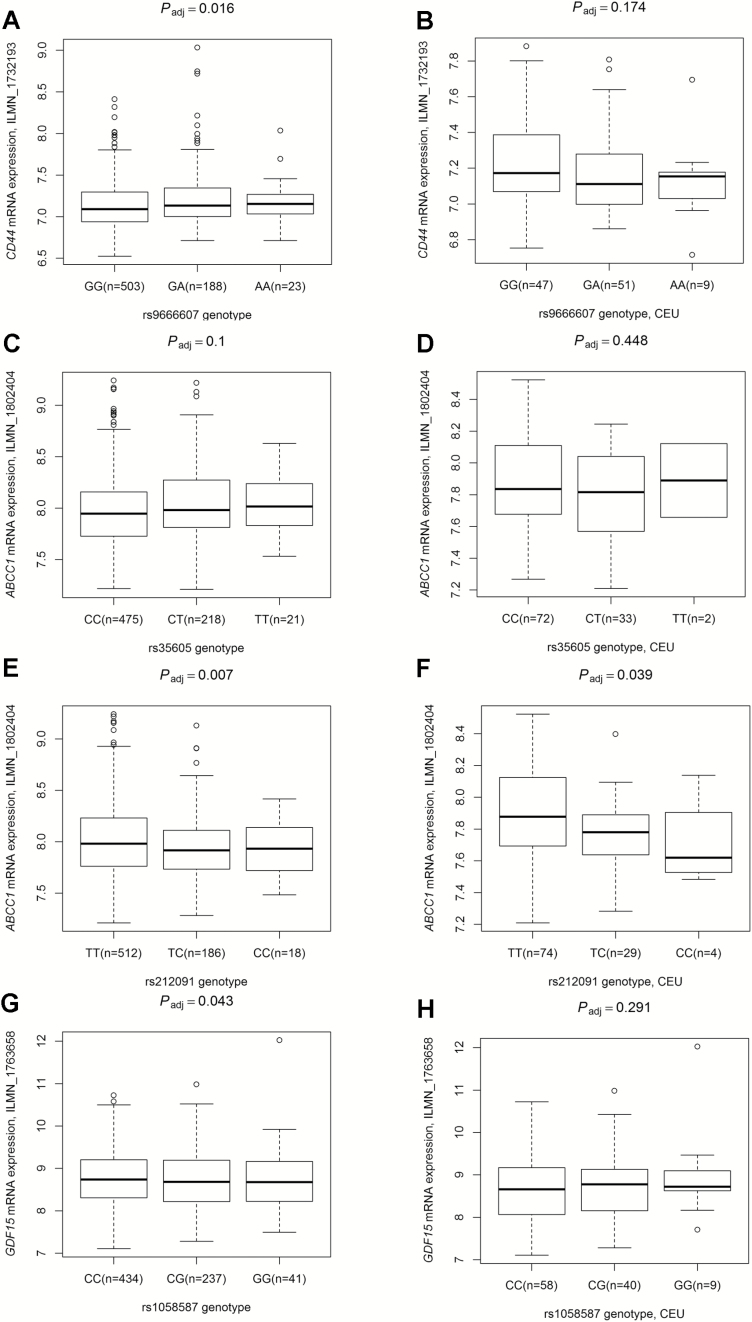
Four independent SNPs showed some evidence of functional relevance using online prediction tools, including SNPinfo and RegulomeDB. Both rs35605 of ABCC1 and rs9666607 of CD44 are located in exonic regions and were predicted to play a role in RNA splicing regulation by SNPinfo. Another SNP of ABCC1 is located in the 3′UTR and was predicted by SNPinfo to affect the miRNA binding site activity. The non-synonymous SNP of GDF15 could result in an amino acid substitution of the corresponding protein product. To provide biological support for the observed associations and predictions, we evaluated the correlation between genotypes of the four independent SNPs and their related mRNA expression levels using mRNA expression data from Epstein–Barr virus-transformed lymphoblastoid cell lines from 716 unrelated individuals in the HapMap3 Project. In the HapMap3 Project, only sex and racial ethnicity were available for each individual. We recalculated the correlation of SNPs and mRNA expression with and without the adjustments in all 716 individuals and 107 CEUs. All results are summarized in Supplementary Table S10, available at Carcinogenesis Online, and Figure 3 was revised with the adjusted P value. ABCC1 rs212091 T>C, GDF15 rs1058587 C>G, and CD44 rs9666607 G>A were significantly associated with mRNA expression levels in overall populations (all Padj < 0.05, Figure 3), of which only the risk A allele of CD44 rs9666607 was associated with a higher mRNA (β = 0.053, Padj = 0.016, Figure 3A). The effect alleles of the other two SNPs (rs212091C and rs1058587G) were all associated with a lower mRNA expression of the corresponding gene (all β < 0, Figure 3E and G). Consistent with overall populations, the analyses in CEU population showed that the effect alleles of ABCC1 rs212091 T>C were significantly associated with lower ABCC1 mRNA expression (Padj < 0.05, β < 0, Figure 3F). In addition, we found that the minor allele T of ABCC1 rs35605 was associated with a higher mRNA expression level (P = 0.018) in prostate tissue from the GTEx Project (http://www.gtexportal.org/) (Supplementary Table S11, available at Carcinogenesis Online).
Figure 3.
Correlations between the four independent and significant SNPs and the relative mRNA expression levels in lymphoblastoid cell lines of 716 individuals from HapMap 3 Project including 107 CEU, 242 CHB, 41 MXL and 326 YRI populations. (A), (C), (E) and (G) are in all populations with 716 individuals. (B), (D), (F) and (H) are only in 107 CEU individuals. (A, B) rs9666607 and CD44 (reporter: ILMN_1732193); (C, D) rs35605 and ABCC1 (reporter: ILMN_1802404); (E, F) rs212091 and ABCC1 (reporter: ILMN_1802404); (G, H) rs1058587 and GDF15 (reporter: ILMN_1763658). Padj is from linear regression analysis adjusted for age and/or ethnic groups. CEU, Utah residents with Northern and Western European ancestry from the CEPH collection; Han, Chinese in Beijing, China; MXL, Mexican ancestry in Los Angeles, CA, USA; YRI, Yoruba in Ibadan, Nigeria.
Discussion
In the present study, we examined whether SNPs of stemness-related genes are associated with PCa survival using available genotyping data from the PLCO. After adjusting for age, Gleason score, stage and primary treatment, we identified four independent SNPs of the CD44, ABCC1 and GDF15 genes that are predicted to be functional and associated with PCa survival. In addition to the role of CD44, ABCC1 and GDF15 in stemness, these genes play roles in tumor cell biology. Specifically, CD44, which encodes cluster of differentiation 44, is a transmembrane glycoprotein (37). In the context of tumor cell biology, an increase in CD44 expression has been associated with metastasis and prognosis. Through its ligands, CD44 mediates cellular adhesion, migration, innate immunity, wound healing, cancer progression, metastasis and activation of oncogenic signaling and transporters. Activation of oncogenic signaling and transporters mediates cellular proliferation, migration, invasion, survival and therapeutic resistance. Consistent with the roles of CD44, the CD44 rs9666607 G/A variant was associated with PCa survival in the present study. There is much evidence that the CD44 gene undergoes alternative RNA splicing. The expression of CD44 in normal and cancer cells and CD44-mediated biological processes has been attributed to distinct CD44 isoforms. In the context of PCa, androgens and the androgen receptor have been shown to play a role in regulating alternative RNA splicing of the CD44 gene (38). SNPs in cis-acting RNA splicing elements influence alternative RNA splicing. Interestingly, the CD44 rs9666607 G/A variant was located in a predicted exonic splicing enhancer and thus predicted to play a role in RNA splicing regulation.
ABCC1, which encodes the ATP-binding cassette, subfamily C, member 1, is a member of the ABC transporter family (39). These transmembrane proteins play an important role in ATP-dependent transport of lipids, metabolites and drugs. ABC transporters have garnered attention in oncology, as overexpression of such proteins, which efflux chemotherapeutic drugs from cells, has been shown to cause multidrug resistance. In the context of PCa, overexpression of ABCC1 has been shown (40). In addition, CD44+/CD133+ PCa cells exhibiting an increased resistance to cisplatin have been isolated, and the knockdown of increased expression of Notch1 in such cells has been shown to decrease expression of ABCC1 and increase sensitivity to cisplatin (41). More recently, ABC transporters are garnering attention with respect to roles in cancer initiation and progression and transport of lipids that effect oncogenic signaling. Substrates of ABCC1 include prostaglandins, leukotrienes and sphingosine-1-phosphate. The minor alleles of the two variants of ABCC1, rs35605 C/T and rs212091 T/C were found to be associated with better PCa survival in the present study. These two ABCC1 variants were predicted to affect specific modes of gene regulation, including RNA splicing and miRNA binding. The rs35605 allele correlated with a trend toward increased expression of ABCC1 in blood cells of healthy individuals. On the other hand, the rs212091 allele was correlated with decreased expression of ABCC1 in blood cells of healthy individuals. Multiple ABCC1 RNA splice variants exist in PCa as well as other types of solid tumors according to The Cancer Genome Atlas (TCGA) Research database. The SNP rs212091 is located in exon 31, which includes the 3′UTR, and differential RNA splicing events involving exon 31 are detected in PCa and other types of solid tumors. The SNP rs35605 is located in exon 13, and differential RNA splicing events involving exon 13, while not detected in PCa in TCGA, are detected in other types of solid tumors in TCGA, including cancers of the breast, colon and lung. This event may occur in stages of PCa not included in TCGA. It is possible that rs212091 and rs35605 regulate RNA splicing of ABCC1 and lead to the opposing effects on expression and function.
GDF15, which encodes growth/differentiation factor-15, is a cytokine and member of the transforming growth factor beta (TGFβ) family (42). It has been implicated in stress response, tissue homeostasis and repair, embryonic development, osteogenesis, hematopoiesis and cancer risk and progression. Specific to PCa, elevated expression of GDF15 has been shown in prostatectomy and tumor-adjacent prostate tissues. GDF15 has shown potential as a biomarker for PCa, as increased levels of GDF15 have been detected in serum from metastatic PCa patients, GDF15 serum levels were associated with PCa prognosis, and GDF15 is one of seven genes found to discriminate tumor and control urine. At present, there is conflicting evidence for GDF15 having PCa suppressive and oncogenic activity. In addition, an SNP of GDF15 has been associated with a decreased risk of PCa and an increased risk of death from PCa (43). Consistent with the roles of GDF15 as an oncogene and a prognostic factor for PCa, the GDF15 rs1058587 C/G variant was found to be associated with PCa survival in the present study. This variant was predicted to result in an amino acid change in the cystine-knot cytokine and TGFβ, C-terminal domain of GDF15.
In conclusion, we have identified SNPs of stemness-related genes that are associated with PCa survival and are predicted to have biological functions. An internal validation procedure, utilizing bootstrap sampling and repeated generation of training and testing sets, demonstrated the utility of the genes we identified beyond the usual prognostic variables. Limitations of the present study include those associated with using available data from the PLCO and HapMap3 Project. The genotype data includes only 1150 non-Hispanic white PCa patients and thus prohibited us from identifying SNPs of stemness-related genes and examining their associations with survival of African American PCa patients. In addition, not having access to the target PCa tissues from the participants prevented us from performing gene expression analysis using such tissues. Furthermore, associations were limited to the clinical variables included in the data set. In the future, these results should be confirmed in a larger, prospective study. In addition, associations of these SNPs with PCa survival should be investigated in a racially diverse cohort. Given the higher PCa incidence and mortality in African Americans and the more aggressive biology of African American PCa, understanding the molecular mechanisms underlying PCa in African Americans and developing associated novel approaches for prevention and treatment that will help reduce PCa disparities for African Americans are urgently needed. Moreover, correlations between genotypes of independent SNPs and their related mRNA expression levels should be investigated in prostate tissue. Furthermore, the functional consequences of the SNPs should be assessed in PCa cells. Finally, generation of a cohort with annotated behavioral, social, neighborhood and physiological factors would enable associations of these SNPs with such factors to be evaluated. Our findings that the AUC of the 10-year OS models significantly increased after adding the genetic scores to clinical variables and that in the time-dependent ROC, the cumulative AUC at different time points were greater than the one including only clinical variables suggest that these genetic factors have the potential to serve as novel molecular targets for development of biomarkers of aggressive PCa. Thus, our recent identification of RNA splicing regulatory variants in stemness-related genes that were significantly associated with racial disparities in susceptibility to PCa (19), and our findings in the present study suggest genetic variation that impacts regulation of RNA splicing of genes involved in stemness could be developed as biomarkers of PCa risk and survival. In addition, given the roles of these stemness-related genes in tumor cell biology, these genetic factors also have the potential to serve as novel molecular targets for developmental therapeutics against aggressive PCa.
Supplementary Material
Supplementary date are available at Carcinogenesis online.
Funding
A DoD Prostate Cancer Research Program Health Disparity Research Award(PC131972 to S.R.P. PI, N.H.L. QC and J.A.F. Co-I); a NIH Feasibility Studies to Build Collaborative Partnerships in Cancer Research P20 Award(1P20-CA202925-01A1 to S.R.P. Overall PI and J.A.F. PI of Pilot Project One); a NIH Basic Research in Cancer Health Disparities R01 Award (R01CA220314 to S.R.P. PI and J.A.F. Co-I); the Duke Cancer Institute and the Duke Cancer Institute’s P30 Cancer Center Support Grant (NIH CA014236 to J.A.F., X.L., P.G.M., D.J.G., T.H., Q.W. and S.R.P.).
Conflict of Interest Statement
None declared.
Acknowledgements
We thank participants of the PLCO and the NCI for providing access to data collected by the PLCO. PLCO was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and contracts from the Division of Cancer Prevention, NCI, NIH and DHHS. The authors thank PLCO screening center investigators and staff, and the staff of Information Management Services and Westat. The statements contained herein are solely those of the authors and do not represent or imply concurrence or endorsement by NCI. The authors also acknowledge dbGaP repository for providing the cancer genotyping data set. The accession number for the data set of CGEMS PCa scan is phs000207.v1.p1.
Abbreviations
- AUC
area under curve
- CI
confidence interval
- eQTL
expression quantitative trait loci
- FPRP
false-positive report probability
- GWAS
genome-wide association study
- HR
hazards ratio
- LD
linkage disequilibrium
- OS
overall survival
- PCa
prostate cancer
- PLCO
Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial
- PSA
prostate-specific antigen
- ROC
receiver-operating characteristic
- SNP
single-nucleotide polymorphism
References
- 1. Siegel R.L., et al. (2016) Cancer statistics, 2016. CA Cancer J. Clin., 66, 7–30. [DOI] [PubMed] [Google Scholar]
- 2. National Institutes of Health, National Cancer Institute, Surveillance, Epidemiology, and End Results Program. SEER Stat Fact Sheets: Prostate http://seer.cancer.gov/statfacts/html/prost.html(22 January 2018 date last accessed).
- 3. Crook J., et al. (2013) Prognostic factors for newly diagnosed prostate cancer and their role in treatment selection. Semin. Radiat. Oncol., 23, 165–172. [DOI] [PubMed] [Google Scholar]
- 4. Boström P.J., et al. (2015) Genomic predictors of outcome in prostate cancer. Eur. Urol., 68, 1033–1044. [DOI] [PubMed] [Google Scholar]
- 5. Beck B., et al. (2013) Unravelling cancer stem cell potential. Nat. Rev. Cancer, 13, 727–738. [DOI] [PubMed] [Google Scholar]
- 6. Collins A.T., et al. (2001) Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J. Cell Sci., 114 (Pt 21), 3865–3872. [DOI] [PubMed] [Google Scholar]
- 7. Richardson G.D., et al. (2004) CD133, a novel marker for human prostatic epithelial stem cells. J. Cell Sci., 117 (Pt 16), 3539–3545. [DOI] [PubMed] [Google Scholar]
- 8. Goldstein A.S., et al. (2008) Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc. Natl Acad. Sci. USA, 105, 20882–20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiao J., et al. (2012) Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS One, 7, e42564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hurt E.M., et al. (2008) CD44+ CD24(−) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br. J. Cancer, 98, 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collins A.T., et al. (2005) Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res., 65, 10946–10951. [DOI] [PubMed] [Google Scholar]
- 12. Hao J., et al. (2012) In vitro and in vivo prostate cancer metastasis and chemoresistance can be modulated by expression of either CD44 or CD147. PLoS One, 7, e40716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kyjacova L., et al. (2015) Radiotherapy-induced plasticity of prostate cancer mobilizes stem-like non-adherent, Erk signaling-dependent cells. Cell Death Differ., 22, 898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qin J., et al. (2012) The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell, 10, 556–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mimeault M., et al. (2011) Frequent gene products and molecular pathways altered in prostate cancer- and metastasis-initiating cells and their progenies and novel promising multitargeted therapies. Mol. Med., 17, 949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Al Olama A.A., et al. (2014) A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat. Genet., 46, 1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lange E.M., et al. (2014) Genome-wide association scan for variants associated with early-onset prostate cancer. PLoS One, 9, e93436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amin Al Olama A., et al. (2013) A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum. Mol. Genet., 22, 408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y., et al. (2017) Associations between RNA splicing regulatory variants of stemness-related genes and racial disparities in susceptibility to prostate cancer. Int. J. Cancer, 141, 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knipe D.W., et al. (2014) Genetic variation in prostate-specific antigen-detected prostate cancer and the effect of control selection on genetic association studies. Cancer Epidemiol. Biomarkers Prev., 23, 1356–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim S., et al. (2015) Genetic variants at 1q32.1, 10q11.2 and 19q13.41 are associated with prostate-specific antigen for prostate cancer screening in two Korean population-based cohort studies. Gene, 556, 199–205. [DOI] [PubMed] [Google Scholar]
- 22. Borque A., et al. (2013) Genetic predisposition to early recurrence in clinically localized prostate cancer. BJU Int., 111, 549–558. [DOI] [PubMed] [Google Scholar]
- 23. San Francisco I.F., et al. (2014) Association of RNASEL and 8q24 variants with the presence and aggressiveness of hereditary and sporadic prostate cancer in a Hispanic population. J. Cell. Mol. Med., 18, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reinhardt D., et al. (2014) Prostate cancer risk alleles are associated with prostate cancer volume and prostate size. J. Urol., 191, 1733–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin H.Y., et al. (2013) SNP-SNP interaction network in angiogenesis genes associated with prostate cancer aggressiveness. PLoS One, 8, e59688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berndt S.I., et al. ; African Ancestry Prostate Cancer GWAS Consortium (2015) Two susceptibility loci identified for prostate cancer aggressiveness. Nat. Commun., 6, 6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andriole G.L., et al. ; PLCO Project Team (2012) Prostate cancer screening in the randomized Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial: mortality results after 13 years of follow-up. J. Natl Cancer Inst., 104, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yeager M., et al. (2007) Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet., 39, 645–649. [DOI] [PubMed] [Google Scholar]
- 29. Xu Z., et al. (2009) SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res., 37, W600–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boyle A.P., et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wacholder S., et al. (2004) Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl Cancer Inst., 96, 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Howie B.N., et al. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aulchenko Y.S., et al. (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics, 23, 1294–1296. [DOI] [PubMed] [Google Scholar]
- 34. Akaike H. (1974) A new look at the statistical model identification. IEEE Trans. Automat. Contr., 19, 716–723. [Google Scholar]
- 35. Chambless L.E., et al. (2006) Estimation of time-dependent area under the ROC curve for long-term risk prediction. Stat. Med., 25, 3474–3486. [DOI] [PubMed] [Google Scholar]
- 36. Stranger B.E., et al. (2012) Patterns of cis regulatory variation in diverse human populations. PLoS Genet., 8, e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ponta H., et al. (2003) CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol., 4, 33–45. [DOI] [PubMed] [Google Scholar]
- 38. Clark E.L., et al. (2008) The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res., 68, 7938–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fletcher J.I., et al. (2010) ABC transporters in cancer: more than just drug efflux pumps. Nat. Rev. Cancer, 10, 147–156. [DOI] [PubMed] [Google Scholar]
- 40. Karatas O.F., et al. (2016) The role of ATP-binding cassette transporter genes in the progression of prostate cancer. Prostate, 76, 434–444. [DOI] [PubMed] [Google Scholar]
- 41. Liu C., et al. (2014) NOTCH1 signaling promotes chemoresistance via regulating ABCC1 expression in prostate cancer stem cells. Mol. Cell. Biochem., 393, 265–270. [DOI] [PubMed] [Google Scholar]
- 42. Vaňhara P., et al. (2012) Growth/differentiation factor-15: prostate cancer suppressor or promoter?Prostate Cancer Prostatic Dis., 15, 320–328. [DOI] [PubMed] [Google Scholar]
- 43. Hayes V.M., et al. (2006) Macrophage inhibitory cytokine-1 H6D polymorphism, prostate cancer risk, and survival. Cancer Epidemiol. Biomarkers Prev., 15, 1223–1225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.