Fig. 1.
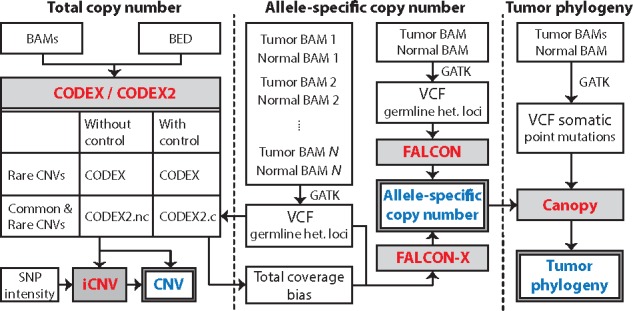
A flowchart outlining the procedures for profiling CNV, ASCN and reconstructing tumor phylogeny. CNVs with common and rare population frequencies can be profiled by CODEX and CODEX2, with and without negative control samples. iCNV integrates sequencing and microarray data for CNV detection. ASCNs can be profiled by FALCON and FALCON-X using allelic read counts at germline heterozygous loci. Canopy infers tumor phylogeny using somatic SNVs and ASCNs
