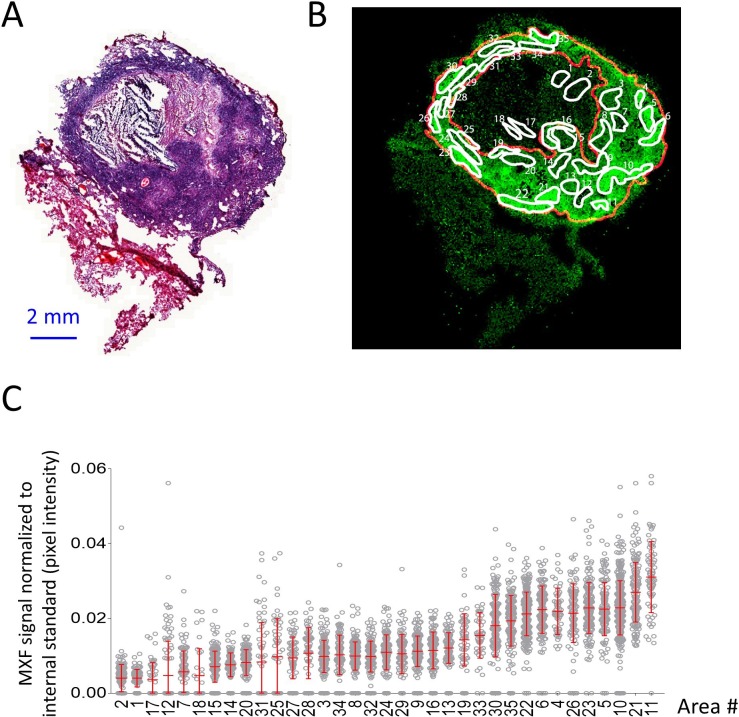
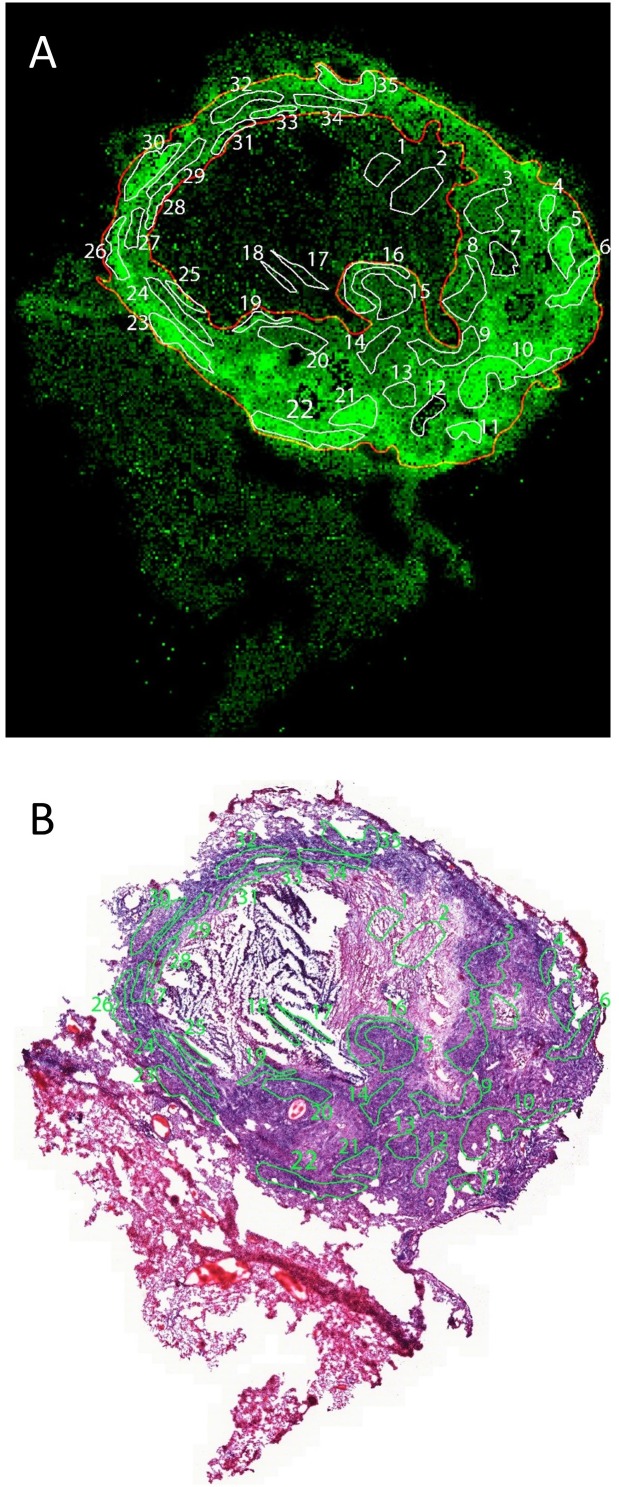
(
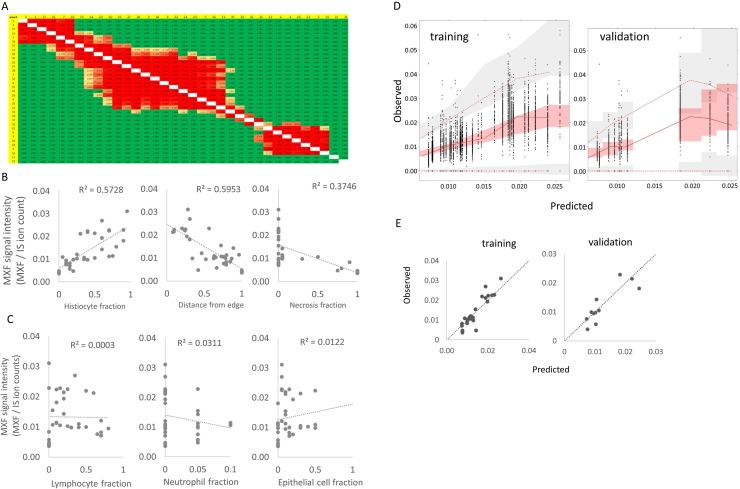
A) Statistical analysis of MXF relative abundance per area: average pixel intensities in each ROI were compared using a one-way analysis of variance (ANOVA) comparing the mean MXF pixel intensity in each of the 35 areas, and a Tukey post-hoc test for multiple comparisons. The number of pixels per ROI ranged from 48 and 429. Green cells indicate comparisons with p≤0.05. (
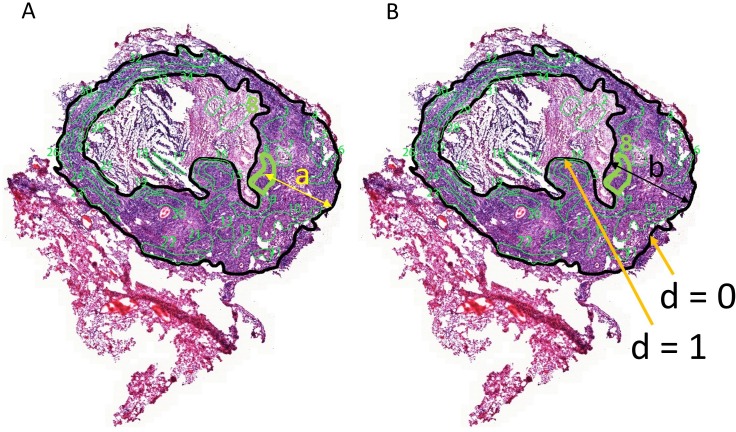
B) Plots of the Spearman rank correlations between MXF abundance and the fraction of histiocytes (including macrophages and foamy macrophages), relative distance of ROI to granuloma edge and fraction of necrosis. The method used to measure the relative distance of each sub-area to granuloma edge, a surrogate of vascular efficiency, is described in
Figure 3—figure supplement 3. (
C) Plots of Spearman rank correlations between MXF abundance and fraction of lymphocytes, neutrophils, and epithelial cells. (
D) Visual Predictive checks of the final model developed on the training dataset (left), and internal validation showing predicted MXF abundance using ROI-specific measurements of histiocytes, distance ratio and % necrotic cells in the final model equation.
where
are model estimated constants and are reported in
supplementary file 1 The lines and shaded areas in D are as follows: continuous red line: observed data median; dotted red line: 5
th and 95
th percentile of the observed data; light red shaded area: predicted median with uncertainty; light grey shaded area: predicted percentiles of the data with uncertainty. (
E) Visual Predictive Checks (VPC) similar to the plots described in (
D) but showing only the median signal intensity for each ROI. Dotted line: line of identity.