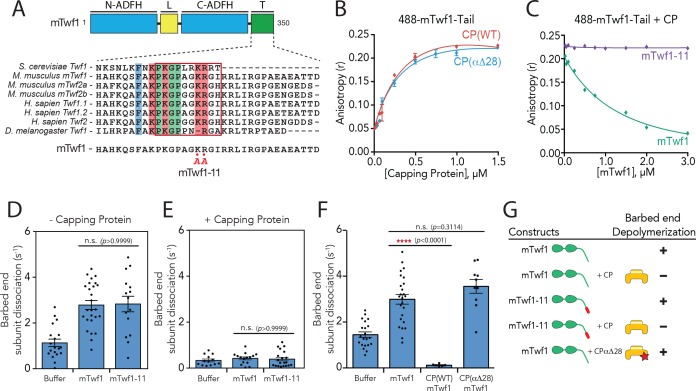
Figure 1. Barbed end capping by Capping Protein inhibits Twinfilin1-mediated depolymerization.
(A) Mouse Twinfilin-1 (mTwf1) domain organization: ADF-H, actin depolymerization factor homology domain; L, linker; T, tail. Sequence alignment of tail regions of Twinfilin isoforms from different species with boxed region highlighting conservation of residues critical for binding to Capping Protein (CP). mTwf1-11 carries a mutation in the tail region (KR332,333AA) that disrupts binding to CP. (B) Fluorescence anisotropy measurement of 100 nM HiLyte488-labeled mTwf1 tail peptide mixed with increasing concentrations of the indicated CP construct. (C) Fluorescence anisotropy measurement of 100 nM HiLyte488-labeled mTwf1 tail peptide incubated in the presence 1 μM CP and increasing concentrations of either mTwf1 or mTwf1-11. Anisotropy values for each condition averaged from three independent experiments. (D,E) Rates of barbed end depolymerization (subunits s−1) induced by 1 μM of the indicated mouse Twinfilin, in the (D) absence or (E) presence of 10 nM CP, determined from TIRF assays. Rates for each condition averaged from at least five filaments in each of two independent experiments. From left to right: (D) n = 19, 26, and 15 and mean depolymerization rates 1.13, 2.784 and 2.81 subunits s−1; (E) n = 13, 15, and 20 and mean depolymerization rates 1.13, 2.784 and 2.81 subunits s−1. (F) Rates of barbed end depolymerization (subunits s−1) induced by 1 μM mTwf1, in the absence or presence of 1 μM of the indicated CP construct, determined from TIRF assays. Rates for each condition averaged from at least five filaments from at least one experiment. From left to right n = 21, 25, 6, and 10; mean depolymerization rates 1.45, 2.991, 0.11, and 3.58 subunits s−1. (G) Summary of barbed end depolymerization activity of mTwf1 constructs in combination with different CP constructs determined from TIRF assays (as in D,E,F). Error bars, s.e.m. ****p≤0.0001, n.s. p>0.05 by one-way ANOVA with Tukey post hoc test.