Figure 5.
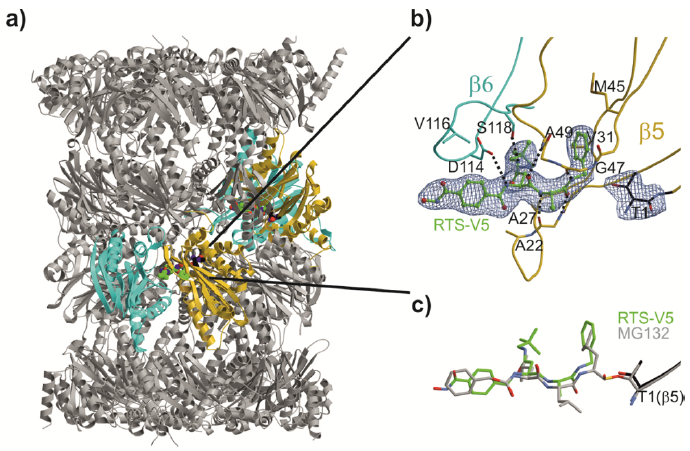
Yeast 20S proteasome in complex with RTS-V5. a) Cartoon representation of the yeast 20S proteasome core particle (yCP) in complex with RTS-V5 (PDB ID: 6H39). The decarboxylated ligand is presented as a sphere model, which is located at the intersection of the β–β′-rings. The molecule solely binds to the nonprimed substrate binding channel of the chymotrypsin-like active site, which is composed of subunits β5 (gold) and β6 (cyan), respectively. b) The 2FO-FC electron density map of the non-covalent inhibitor is illustrated as blue mesh and contoured to 1σ. Hydrogen bonds forming the antiparallel β-sheet between ligand and protein main chain residues are indicated by black dashed lines. RTS-V5 intensely interacts with the S1 and S3 sites, whereas the P2-Ala side chain is solvent-exposed. Amino acid numbering is according to Löwe et al.18 and Groll et al.19. c) Structural superposition of RTS-V5 with the covalently acting aldehyde inhibitor MG132 (PDB ID: 4NNN)20 depicts a uniform arrangement. The hemiacetal bond is highlighted in gold.
