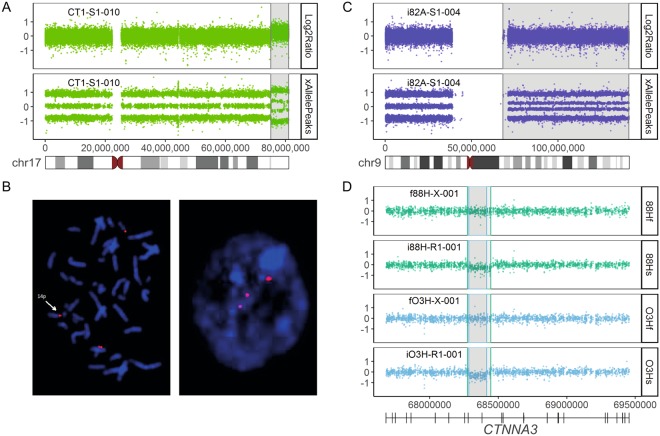
Figure 3.
Examples of CNVs detected by SNP-based CMA. (A) Copy number analysis identified a chromosome 17q terminal gain not detectable with conventional karyotyping in the SiPSC line “CT1-S1-010”. (B) FISH analysis showing the unbalanced translocation 14p/17q in this clone (left = metaphase, right = interphase). (C) Conventional karyotyping and copy number analysis of chromosome 9 of the SiPSC line “i82A-S1-004” revealed unremarkable results (Log2Ratio top), but SNP allele peak distribution (xAllelePeaks bottom) uncovered a copy neutral allelic imbalance on the long arm of chromosome 9 (4 bands) while the short arm (left) shows normal allelic distribution (3 bands) (see also Fig. S2). (D) Two independent overlapping intragenic deletions in the CTNNA3 gene detected in the RiPSC lines “i88H-R1-001” (green bottom) and “iO3H-R1-001” (blue bottom) and absent from their fibroblast cultures “f88H-X-001” (green top) and “iO3H-X-001” (blue top).