Abstract
The Ebola virus-encoded major matrix protein VP40 traffics to the plasma membrane, which leads to the formation of filamentous viral particles and subsequent viral egress. However, the cellular machineries underlying this process are not fully understood. In the present study, we have assessed the role of host endocytic recycling in Ebola virus particle formation. We found that a small GTPase Rab11, which regulates recycling of molecules among the trans-Golgi network, recycling endosomes, and the plasma membrane, was incorporated in Ebola virus-like particles. Although Rab11 predominantly localized in the perinuclear region, it distributed diffusely in the cytoplasm and partly localized in the periphery of the cells transiently expressing VP40. In contrast, Rab11 exhibited a perinuclear distribution when 2 VP40 derivatives that lack ability to traffic to the plasma membrane were expressed. Finally, expression of a dominant-negative form of Rab11 or knockdown of Rab11 inhibited both VP40-induced clusters at the plasma membrane and release of viral-like particles. Taken together, our findings demonstrate that Ebola virus exploits host endocytic recycling machinery to facilitate the trafficking of VP40 to the cell surface and the subsequent release of viral-like particles for its establishment of efficient viral egress.
Keywords: Ebola virus, Rab11, recycling endosome, viral-like particles, virus egress, VP40
Ebola virus (EBOV), a member of the family Filoviridae, is an enveloped, single-stranded, negative-sense ribonucleic acid (RNA) virus that causes severe hemorrhagic fever with a high mortality rate in humans and nonhuman primates [1]. Currently, there are no US Food and Drug Administration-approved therapeutics to treat EBOV infection [2]. Ebola virus encodes 7 structural genes that assemble to yield distinct filamentous viral particles. The EBOV-encoded major matrix protein VP40 traffics to the plasma membrane (PM), which leads to the formation and release of the virus-like particles (VLPs), even when expressed alone [3–7]. VP40 has been shown to form a dimer that further assembles into a flexible filamentous matrix structure [8].
VP40 contains 2 overlapping late domains (PTAP and PPXY motifs) that interact with Tsg101, a component of the endosomal sorting complex required for transport (ESCRT)-1, and with the ubiquitin ligase Nedd4, respectively [9]. Although both late domains are responsible for release of VLPs [10, 11], a study using recombinant EBOV showed that the late domains are dispensable for virus replication [12]. Results from previous studies showed that several host factors are required for the intracellular transport of VP40, including actin [13–15], IQGAP1, a ubiquitously expressed scaffolding protein that regulates processes that include cell motility and division, actin polymerization, and formation of filopodia [16], Sec24C, a component of the host COPII vesicular transport system [17], microtubules [18], and the HECT family E3 ubiquitin ligase WWP1 [19]. However, the molecular mechanisms of VP40-mediated EBOV egress have not been fully elucidated.
The host endocytic recycling machinery and its major regulator, a small GTPase Ras-related in brain (Rab) 11, are implicated in the virus lifecycle including assembly and viral egress [20]. Rab11 associates with recycling endosomes and regulates recycling processes of proteins and vesicles to the cell surface [21]. Rab11 also localizes to the trans-Golgi network (TGN) and post-Golgi vesicles and has been implicated in the trafficking between the TGN and the endosomal recycling compartments through the regulated secretion pathway [22].
The Rab11-dependent recycling pathway involves trafficking of virus components to their sites of egress. Results from previous studies showed that Rab11-positive vesicles associate with viral ribonucleoproteins of a variety of viruses, including influenza A virus (IAV) [23–26], Sendai virus [27, 28], and human parainfluenza virus type I [28], and promote their trafficking toward their sites of egress. Herpes simplex virus-1 exploits the same pathway to transport its viral glycoproteins on the PM to the intracellular compartment for maturation of virions [29]. The Nipah virus-encoded fusion protein is activated by cleavage with cathepsin B in the recycling endosomes and subsequently recycled to the PM for incorporation in the virions [30]. The accessory protein Vpu of human immunodeficiency virus 1 is transported via Rab11-positive vesicles toward the assembly site on the PM [31].
Moreover, a potential role for the Rab11-dependent endocytic recycling pathway in viral egress has been suggested for both positive- and negative-sense RNA viruses. The Rab11-dependent pathway involves budding of respiratory syncytial virus (RSV) [32, 33] and the filamentous IAV [34] by facilitating fission of viral particles from the PM. The Rab11-mediated pathway is also implicated in egress from the PM for Mason-Pfizer monkey virus [35, 36], Jaagsiekte sheep retrovirus [37], and hepatitis C virus [38], which are originally assembled and mature, respectively, at the microtubule organizing center, the endoplasmic reticulum, and the Golgi/TGN compartments.
Although a previous study using liquid chromatography-linked tandem mass spectroscopy (LC-MS/MS) showed that Rab11b is incorporated into authentic filovirus virions [39], its physiological relevance in the EBOV lifecycle has remained unclear. In the present study, we have assessed the role of the Rab11-dependent recycling pathway in VP40-mediated budding of Ebola VLPs. We found that Rab11 was incorporated into Ebola VLPs. Although Rab11 predominantly localized in the perinuclear region, it distributed diffusely in the cytoplasm and partly localized in the periphery of the cells transiently expressing VP40. Blocking of endosomal recycling by the expression of a dominant-negative form of Rab11 or by knockdown of endogenous Rab11 suppressed the VP40-induced formation of clusters at the PM. Moreover, downregulation of Rab11 reduced the production of Ebola VLPs. Taken together, our findings indicate that EBOV exploits host endocytic recycling machinery for its establishment of efficient viral egress.
MATERIALS AND METHODS
Cell Culture
African green monkey kidney epithelial Vero-E6 cells and human embryonic kidney HEK293T cells (American Type Culture Collection) were grown in high-glucose Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and antibiotics. Cells were maintained at 37°C in 5% CO2. Transfection of Vero-E6 and HEK293T cells with expression plasmids and small interfering RNA (siRNA) was carried out with TransIT-LT1 (Mirus, Madison, WI) and TransIT-TKO (Mirus), respectively.
Plasmids
The expression plasmids of EBOV VP40 L117R and I307R mutants were constructed by insertion into the pCAGGS plasmid at the EcoRI and BglII sites. VP40 mutant complementary deoxyribonucleic acids were amplified by polymerase chain reaction using the following oligonucleotides: VP40-L117R, forward 5’-actacggccgccatcatgcgtgcttcatac-3’ and reverse 5’-gggtgatagtgtatgaagcacgcatgatgg-3’; VP40-I307R, forward 5’-ggagacctcaccatggtaagaacacaggatt-3’ and reverse 5’-acgtgtcacaatcctgtgttcttaccatggt-3’. The pEGFP-C3 plasmids encoding green fluorescent protein (GFP)-fused wild-type Rab11 (GFP-wtRab11) and a dominant-negative form of Rab11 (GFP-dnRab1) were kind gifts from Dr. Angela Wandinger-Ness (University of New Mexico) [40].
Preparation of Ebola Virus-Like Particles
The preparation of Ebola VLPs was described previously [41, 42]. In brief, equal amounts of the pCAGGS expression plasmids for EBOV subtype Zaire, strain Mayinga VP40, glycoproteins (GP), and nucleoproteins (NP) were transfected into HEK293T cells. Forty-eight hours posttransfection (h.p.t.), the culture supernatants were harvested and centrifuged at 1500 rpm for 5 minutes and then at 3500 rpm for 15 minutes to remove detached cells and cell debris, respectively. The VLPs were pelleted through a 30% sucrose cushion by centrifugation at 11000 rpm for 1 hour at 4°C with an SW40 rotor (Beckman, Fullerton, CA). The pelleted VLPs were resuspended in phosphate-buffered saline (PBS). For a protease protection assay, Ebola VLPs were treated with or without 0.1 mg/mL trypsin in the presence or absence of 0.05% Triton X-100 at room temperature for 30 minutes, followed by the addition of sodium dodecyl sulfate polyacrylamide gel electrophoresis sample buffer. The incorporation of VP40 and Rab11 in the purified VLPs was confirmed by Western blotting with the mouse monoclonal antibodies against VP40 (clone 6; 1:4000 dilution) and Rab11 (1:1000 dilution; Becton, Dickinson and Company, Franklin Lakes, NJ), respectively. The experiment was performed 3 times independently, and the intensity of the bands was quantified by use of Multigauge software (FUJIFILM Corporation).
Immunofluorescence Staining
Vero-E6 cells grown on coverslips were transfected with the expression plasmid for VP40, VP40-L117R, or VP40-I307R. At 48 h.p.t., the cells were fixed with 4% paraformaldehyde in PBS for 10 minutes at room temperature, permeabilized with PBS containing 0.05% Triton X-100 for 10 minutes at room temperature, and blocked in PBS containing 4% bovine serum albumin for 20 minutes at room temperature. The cells were then incubated with a mouse monoclonal antibody against VP40 (clone 6; 1:2000 dilution) or a rabbit monoclonal antibody against Rab11 (1:200 dilution; Abcam, Cambridge, UK) for 1 hour at room temperature. The cells were then washed 3 times in PBS and incubated with Alexa Fluor 488 or 594-labeled secondary antibodies (1:2000 dilution; Thermo Fisher Scientific, Waltham, MA) for 1 hour at room temperature After washing, nuclei were counterstained with Hoechst 33342 (Cell Signaling Technology, Danvers, MA). To determine the effect of expression of GFP-wtRab11 or -dnRab11 on the distribution of VP40, Vero-E6 cells were transfected with pEGFP-C3 plasmids encoding GFP-wtRab11 or -dnRab11. Forty-eight hours later, the cells were transfected with the expression plasmid for VP40. At 48 h.p.t., the cells were harvested followed by immunofluorescence staining with the antibody for VP40. Images were collected with the 60× oil-immersion objective lens of a confocal laser scanning microscope (Fluoview FV10i; Olympus, Tokyo, Japan) and acquired by using FV10-ASW software (Olympus). Line scan imaging was performed by using FV10-ASW software.
Small Interfering Ribonucleic Acid Treatment
Target sequences corresponding to the human Rab11a and Rab11b-coding sequences were selected and synthesized (Life Technologies) [43]. As a control, an siRNA encoding a sequence that does not target any known genes (Thermo Fisher Scientific) was used. The siRNAs against Rab11a and/or Rab11b were transfected into Vero-E6 or HEK293T cells. At 72 h.p.t, the downregulation of Rab11 in Vero-E6 and HEK293T cells was analyzed by Western blotting and immunofluorescence staining with a mouse anti-Rab11 monoclonal antibody (Becton, Dickinson and Company), a rabbit anti-Rab11 monoclonal antibody (Abcam), and a rabbit anti-β-actin polyclonal antibody (MBL), respectively. For the analysis of the effect of downregulation of Rab11 on distribution of VP40, siRNA-transfected Vero-E6 cells were then transfected with the expression plasmid of VP40 at 72 h.p.t., followed by immunofluorescence staining by means of a mouse monoclonal antibody for VP40. For the analysis of the effect of downregulation of Rab11 on production of Ebola VLPs, siRNA-transfected HEK293T cells were then transfected with the expression plasmids of VP40, NP, and GP at 72 h.p.t., followed by purification of Ebola VLPs as described above. Expression of EBOV proteins in the total cell lysates and the purified VLPs was confirmed by Western blotting with the mouse monoclonal antibodies against VP40 (clone 6; 1:4000 dilution), NP (clone 7.42.18; 1:4000 dilution), or GP (clone 133.13.16.; 1:4000 dilution), respectively. The experiment was performed 3 times independently, and the intensity of the bands was quantified by use of Multigauge software.
RESULTS
Rab11 Is Incorporated Into Ebola Virus-Like Particles
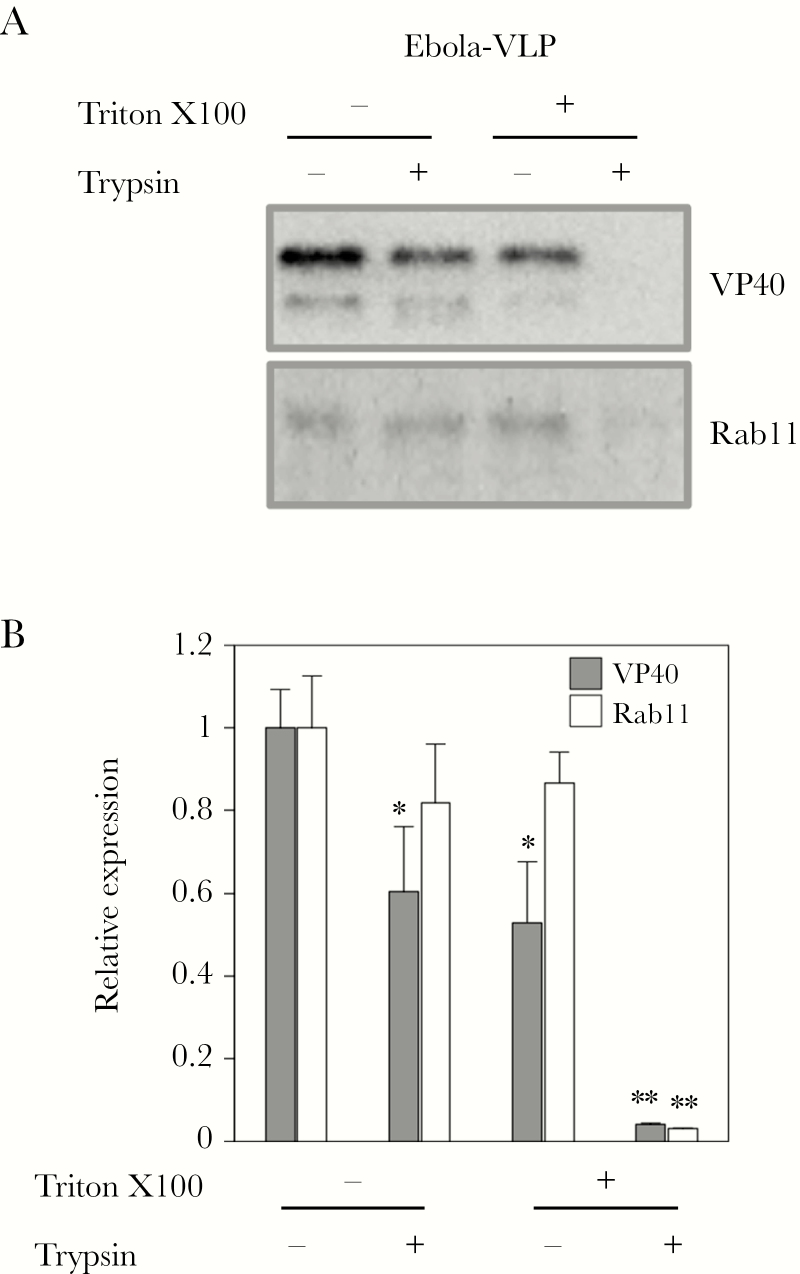
In the present study, we first examined whether Rab11 is incorporated into Ebola VLPs by a protease protection assay [5]. We generated Ebola VLPs by coexpressing VP40, GP, and NP in HEK293T cells [41, 42]. Ebola VLPs were treated with or without trypsin in the presence or absence of Triton X-100 at room temperature for 30 minutes followed by Western blot analysis. Although both VP40 and Rab11 were detected in Ebola VLPs treated with trypsin or Triton X-100 alone, trypsinization in the presence of Triton X-100 significantly abolished the signals of VP40 and Rab11 (Figure 1), indicating the incorporation of Rab11 in Ebola VLPs.
Figure 1.
Characterization of incorporation of Rab11 in Ebola virus-like particles (VLPs). HEK293T cells were transfected with expression plasmids for Ebola virus VP40, glycoproteins, and nucleoproteins. At 48 hours posttransfection, culture medium was harvested. Viral particles were purified from the culture medium and treated with or without trypsin in the presence or absence of Triton X-100. Incorporation of VP40 and Rab11 in Ebola VLPs was analyzed by Western blot analysis. The experiment was performed 3 times independently, and a representative blot is shown (A). The intensity of the bands was quantified, and the average of the relative expression values and its standard deviation are shown (B). *, P < .05; **, P < .01 vs respective control (Student’s t test).
VP40 Induces a Dispersed Distribution of Rab11
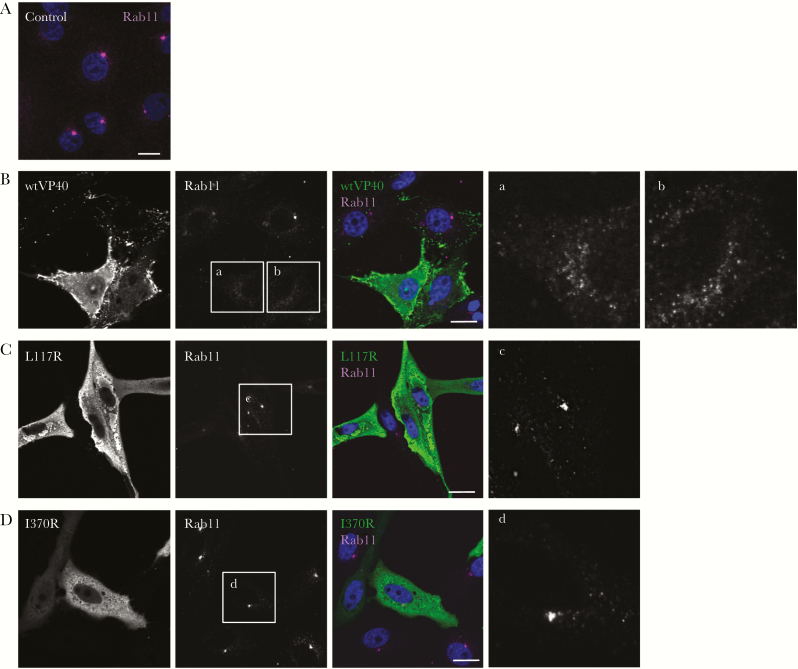
We then investigated the subcellular localization of Rab11 in Vero-E6 cells transiently expressing the EBOV VP40 by immunofluorescence staining. Rab11 predominantly distributed in perinuclear regions, and some fraction of the protein distributed in the cytoplasm in backbone plasmid-transfected control cells (Figure 2A), which is consistent with a prior study [40] showing its localization to recycling endosomes and TGN. In agreement with previous findings [4, 5, 44–46], VP40 distributed in multiple subcellular compartments (Figure 2B). VP40 was visualized diffusely in the cytoplasm and the nucleus, and particularly in the PM as intense clusters. In contrast, co-immunofluorescence staining revealed the disperse distribution of Rab11 throughout the cytoplasm in the cells expressing high levels of VP40 (Figure 2B, insets a and b). However, no efficient colocalization of Rab11 along with VP40 was observed.
Figure 2.
The subcellular distribution of Rab11 in Vero-E6 cells transiently expressing VP40 derivatives. Vero-E6 cells grown on coverslips were transfected with the expression plasmids for wild-type VP40 (B), VP40 L117R (C), or VP40 I370R (D). At 48 hours posttransfection, the cells were harvested and the subcellular distribution of VP40 and Rab11 was analyzed by immunofluorescence staining. As a control, a backbone plasmid was transfected (A). In merged images, VP40 and Rab11 are shown in green and magenta, respectively. The nuclei (blue) were counterstained with Hoechst 33342. Insets show the boxed areas. Scale bars = 10 µm.
We also analyzed the distribution of Rab11 in cells transiently expressing 2 types of VP40 derivatives, L117R (Figure 2C) and I370R (Figure 2D), which lack the ability to traffic to the PM and subsequent VLP formation [8]. In agreement with previous findings, both mutants remained in the cytoplasm and failed to form clusters at the PM. Rab11 distributed in the perinuclear region of cells expressing both VP40 mutants, suggesting its dispersed distribution is likely associated with trafficking of VP40 to the PM.
The Effect of a Dominant-Negative Form of Rab11 on Distribution of VP40
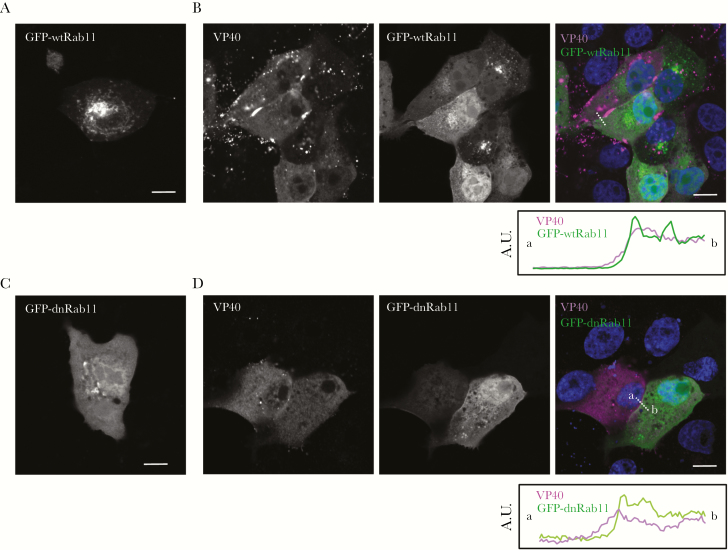
Because the distribution of Rab11 was affected by VP40 (Figure 2), we investigated the role of Rab11 in VP40-induced viral particle formation. We transfected Vero-E6 cells and transiently expressed both GFP-wtRab11 or a dominant-negative form of Rab11 (GFP-dnRab11), which has an amino acid substitution (S25N) [40], and expression plasmids for VP40. We then assessed the effect of Rab11 derivatives on the distribution of VP40 by immunofluorescence staining. When GFP-wtRab11 was expressed alone, it exhibited a predominant distribution in the perinuclear region in the same way as endogenous Rab11 (Figure 2A vs Figure 3A). In contrast, GFP-wtRab11 was predominantly distributed diffusely throughout the cytoplasm in VP40-positive cells (Figure 3B). VP40 distributed in the periphery of the cells expressing GFP-wtRab11 with intense clusters (Figure 3B). GFP-dnRab11 predominantly distributed in the cytoplasm in both VP40-negative and VP40-positive cells (Figure 3C and 3D). In the presence of GFP-dnRab11 expression, VP40 distributed in the cytoplasm without formation of clusters in the PM, suggesting that the expression of GFP-dnRab11 suppressed formation of VP40-induced clusters in the PM (Figure 3D). GFP-wtRab11 and GFP-dnRab11 were partly distributed in the PM of the cells expressing VP40 (Figure 3B and 3D). However, efficient colocalization of Rab11 derivatives with VP40 was not observed, which is consistent with the result in Figure 2B.
Figure 3.
The effect of a dominant-negative form of Rab11 on the distribution of VP40. Vero-E6 cells grown on coverslips were transfected with the expression plasmids for green fluorescent protein (GFP)-wtRab11 (B) or -dnRab11 (D). At 48 hours posttransfection (h.p.t.), the cells were transfected with the expression plasmids for VP40. At 48 h.p.t., the cells were harvested and the subcellular distribution of VP40 was analyzed by immunofluorescence staining. As a control, GFP-wtRab11 (A) or -dnRab11 (C) was expressed alone. In merged images, VP40 and GFP-fused Rab11 derivatives are shown in magenta and green, respectively. The nuclei (blue) were counterstained with Hoechst 33342. Scale bars = 10 µm. The plots indicate the individual fluorescence intensity along each of the corresponding lines. Abbreviation: A.U., arbitrary unit.
A Rab11-Dependent Endocytic Recycling Pathway Contributes to VP40-Mediated Ebola Virus-Like Particle Formation
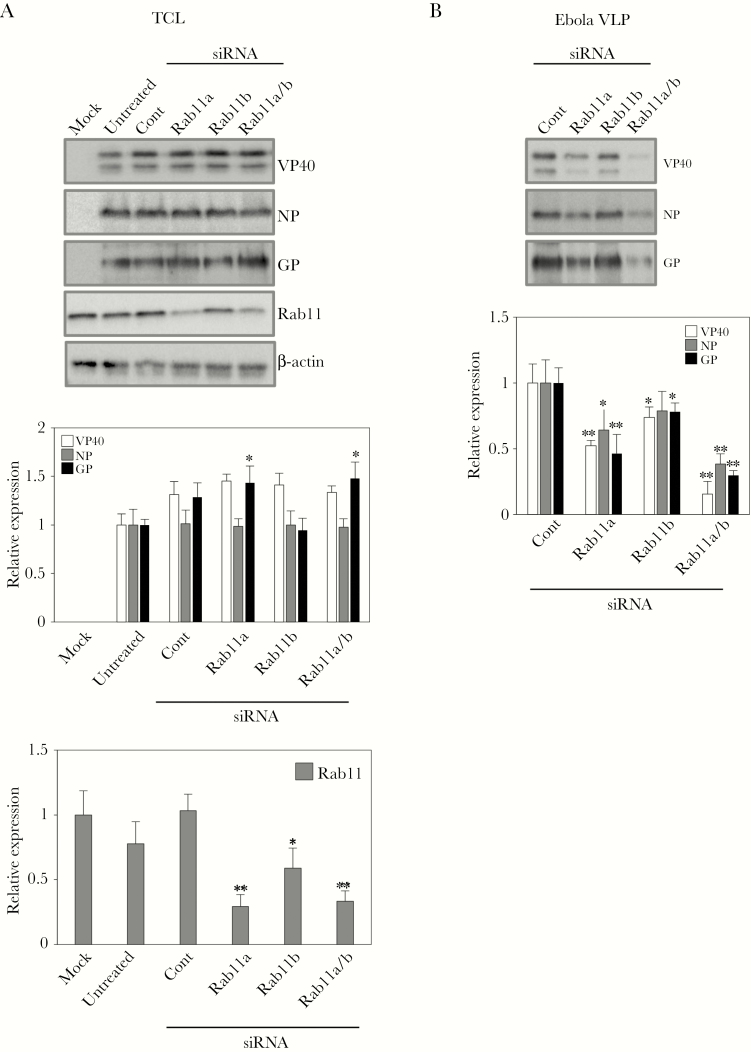
We further assessed the role of Rab11 in VP40-mediated viral particle formation by knockdown of endogenous Rab11 isoforms by means of siRNAs. Rab11 consists of 3 isoforms: Rab11a, Rab11b, and Rab25. Rab11a and Rab11b share 90% amino acid sequence identity, whereas Rab11a and Rab11b share approximately 60% identity with Rab25. Rab11a is expressed ubiquitously, predominantly localizes to recycling endosomes, and functions in the recycling of a wide range of molecules to the cell surface [22]. Rab11b is expressed in the heart, brain, and testes and functions in recycling of molecules in polarized cells [47]. Rab25 has been shown to contribute to the invasiveness of cancer cells by promoting integrin trafficking [48]. Multiple studies have demonstrated that Rab11a functions in the lifecycle of a variety of viruses [20]. Moreover, LC-MS/MS has demonstrated the incorporation of Rab11b into authentic filovirus virions [39]. Thus, we assessed the effect of downregulation of Rab11a and Rab11b isoforms in VP40-mediated formation of Ebola VLPs. We transfected HEK293T cells with siRNAs for Rab11a and/or Rab11b isoforms. At 72 h.p.t, expression plasmids for VP40, NP, and GP were transfected to produce VLPs. We confirmed that knockdown of Rab11 isoforms minimally affected the expression of VP40 (Figure 4A). We found that the downregulation of both Rab11a alone and of both its isoforms significantly suppressed the formation of Ebola VLPs (Figure 4B), indicating that Rab11 plays a crucial role in the budding of Ebola viral particles.
Figure 4.
The role of Rab11 in budding of Ebola virus-like particles (VLPs). HEK293T cells were transfected with small interfering ribonucleic acid (siRNA) against Rab11a, Rab11b alone, or Rab11a and Rab11b. At 72 hours posttransfection (h.p.t.), the cells were transfected with the expression plasmids for VP40, nucleoproteins (NP), and glycoproteins (GP). At 48 h.p.t., the cells and culture medium were harvested. Viral particles were purified from the culture medium. Total cell lysates (TCL) (A) and VLPs (B) were analyzed by Western blotting with the antibodies against VP40, NP, GP, Rab11, or β-actin. The experiment was performed 3 times independently, and the representative blot is shown. The intensity of the bands correspondence to each protein was quantified. The intensity of corresponding bands to VP40, NP, GP, or Rab11 was normalized with that of corresponding bands to β-actin in the same sample. The average of relative expression values and its its standard deviation are shown. *, P < .05; **, P < .01 vs respective control (Student’s t test).
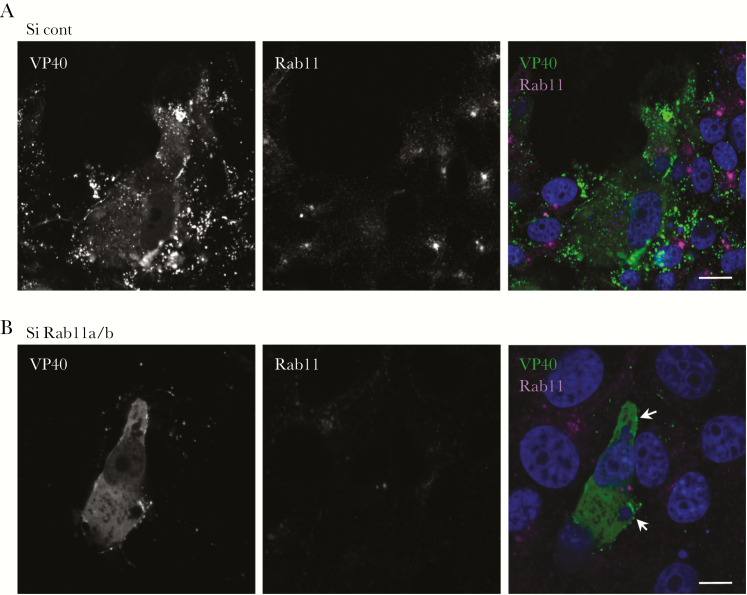
We further examined the effect of Rab11 knockdown on the distribution of VP40 in Vero-E6 cells. The cells in which Rab11a and b isoforms were downregulated suppressed the formation of VP40-positive clusters at the PM (Figure 5B), which was observed in control siRNAs-treated cells (Figure 5A). Remaining clusters on the PM (white arrows) are likely derived from the incomplete knockdown of Rab11 (Figure 4A). The data indicate that the cluster formation of VP40 to the PM and the subsequent VLP formation require Rab11-dependent endocytic recycling pathway.
Figure 5.
The role of Rab11 in VP40-induced clusters in the plasma membrane (PM). Vero-E6 cells were transfected with control small interfering ribonucleic acids (siRNAs) (A) or siRNAs against Rab11a and Rab11b (B). At 72 hours posttransfection (h.p.t.), the cells were transfected with the expression plasmid of VP40. At 48 h.p.t., the subcellular distribution of VP40 and Rab11 was analyzed by immunofluorescence staining. In merged images, VP40 and Rab11 are shown in green and magenta, respectively. The nuclei (blue) were counterstained with Hoechst 33342. White arrows represent the remaining VP40 clusters in the PM. Scale bars = 10 µm.
DISCUSSION
It has been shown that Rab11 is pivotal for both assembly and egress of various viruses [9]. In this study, we demonstrate that Rab11 plays an important role in trafficking of VP40 to the PM and the subsequent budding of Ebola VLPs.
Rab11 is a multifunctional molecule that is involved in the endocytic recycling pathway as well as actin remodeling, cytokinesis, and abscission. Rab11’s diverse functions are regulated by interactions with Rab11 family interacting proteins (Rab11-FIPs), which direct Rab11 to specific subcellular locations by binding to actin- or microtubules-associated motor proteins [49].
We demonstrated that Rab11 distributed more diffusely in the cytoplasm upon expression of VP40 (Figure 2B) and was subsequently incorporated into Ebola VLPs (Figure 1). We also observed that blocking Rab11 function by expression of a dominant-negative form of Rab11 (Figure 3) and downregulation of Rab11 by siRNAs (Figure 5) abrogated the formation of VP40-induced clusters at the periphery of the cells.
These data indicate that VP40 induced an alteration in Rab11-mediated vesicular trafficking toward the PM. Moreover, 2 types of VP40 derivatives, which lack the ability to transport to the PM and subsequent VLP formation, failed to induce the dispersed distribution of Rab11 (Figure 2C and 2D), suggesting that trafficking of VP40 to the PM is likely associated with the Rab11-dependent trafficking machinery.
The Rab11-mediated recycling pathway involves budding of RSV [32, 33] and filamentous IAV virions [34]. For these viruses, Rab11 appears to promote remodeling of membrane morphology, which leads to the scission needed to release particles from the cell surface. The Rab11 effectors proteins FIP1 and FIP2 have been found to facilitate fission of RSV from the apical cell surface. In particular, FIP2 controls the length of the filamentous RSV virions [33]. Moreover, Rab11-FIP3 has been shown to be involved in membrane scission to release filamentous IAV virions from the PM. Rab11-FIP2 is known to (1) recruit the myosin Vb (MyoVb) motor protein for the tethering of recycling endosomes to actin at the microtubule-actin junction and (2) coordinate delivery to the PM [50]. Rab11-FIP3 interacts with the actin-regulating protein Arf6 at the PM [51]. Thus, it is most likely that these Rab11 effector proteins mediate the remodeling of the actin cytoskeleton at the PM, which is responsible for the budding of these viruses.
Although our data indicate that siRNA treatment for Rab11 suppressed the formation of VP40-positive clusters in the PM (Figure 5) and the production of Ebola VLPs (Figure 4), it is still unclear whether the Rab11-mediated recycling pathway is directly responsible for the cluster formation of VP40 in the PM and/or budding of Ebola VLPs from the cell surface. Moreover, fluorescence staining analysis revealed that efficient colocalization of VP40 with endogenous Rab11 and GFP-wtRab11 was not observed (Figures 2 and 3). Further ultrastructural analyses of the morphology of budding VLPs on the cell surface upon blocking Rab11 function will be required to clarify the role of Rab11 in the process of Ebola VLPs formation. In parallel, characterization of the Rab11-associtated effector proteins underlying this process is needed to better understand the detailed molecular mechanism by which Rab11 contributes to the intracellular trafficking of VP40 and formation of Ebola VLPs.
CONCLUSIONS
In summary, our findings indicate that the Rab11-dependent endocytic pathway mediates both trafficking of VP40 to the cell surface and efficient EBOV egress.
Notes
Acknowledgments. We thank to Dr. Angela Wandinger-Ness (University of New Mexico) for providing the pEGFP-C3 plasmids encoding green fluorescent protein-fused Rab11 derivatives. We also thank to Dr. Bill Sugden for critical reviewing of this manuscript.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Financial support. This work was supported by grants from Japan Society for the Promotion of Science (23790493), Japan Science and Technology Agency (01-117), Daiichi Sankyo Foundation of Life Science, Hayashi Memorial Foundation for Female Natural Scientists, Mochida Memorial Foundation for Medical and Pharmaceutical Research, Suhara Memorial Foundation, and Akiyama Life Science Foundation (to A. N.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Feldmann H, Sanchez A, Geisbert T. Filoviridae: Marburg and Ebola viruses. In: Knipe M, Howley PM, eds. Fields Virology. 6th ed. Philadelphia, PA: Lippincott Williams & Wilkins, 2013. [Google Scholar]
- 2. Wong G, Kobinger GP. Backs against the wall: novel and existing strategies used during the 2014-2015 Ebola virus outbreak. Clin Microbiol Rev 2015; 28:593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kolesnikova L, Bamberg S, Berghöfer B, Becker S. The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: exploiting the retrograde late endosomal pathway. J Virol 2004; 78:2382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hoenen T, Biedenkopf N, Zielecki F, et al. Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J Virol 2010; 84:7053–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jasenosky LD, Neumann G, Lukashevich I, Kawaoka Y. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J Virol 2001; 75:5205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Noda T, Watanabe S, Sagara H, Kawaoka Y. Mapping of the VP40-binding regions of the nucleoprotein of Ebola virus. J Virol 2007; 81:3554–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Timmins J, Scianimanico S, Schoehn G, Weissenhorn W. Vesicular release of ebola virus matrix protein VP40. Virology 2001; 283:1–6. [DOI] [PubMed] [Google Scholar]
- 8. Bornholdt ZA, Noda T, Abelson DM, et al. Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell 2013; 154:763–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe 2013; 14:232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci U S A 2000; 97:13871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yasuda J, Nakao M, Kawaoka Y, Shida H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J Virol 2003; 77:9987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neumann G, Ebihara H, Takada A, et al. Ebola virus VP40 late domains are not essential for viral replication in cell culture. J Virol 2005; 79:10300–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adu-Gyamfi E, Digman MA, Gratton E, Stahelin RV. Investigation of Ebola VP40 assembly and oligomerization in live cells using number and brightness analysis. Biophys J 2012; 102:2517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han Z, Harty RN. Packaging of actin into Ebola virus VLPs. Virol J 2005; 2:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kolesnikova L, Ryabchikova E, Shestopalov A, Becker S. Basolateral budding of Marburg virus: VP40 retargets viral glycoprotein GP to the basolateral surface. J Infect Dis 2007; 196(Suppl 2):S232–6. [DOI] [PubMed] [Google Scholar]
- 16. Liu Y, Cocka L, Okumura A, Zhang YA, Sunyer JO, Harty RN. Conserved motifs within Ebola and Marburg virus VP40 proteins are important for stability, localization, and subsequent budding of virus-like particles. J Virol 2010; 84:2294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamayoshi S, Noda T, Ebihara H, et al. Ebola virus matrix protein VP40 uses the COPII transport system for its intracellular transport. Cell Host Microbe 2008; 3:168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noda T, Ebihara H, Muramoto Y, et al. Assembly and budding of Ebolavirus. PLoS Pathog 2006; 2:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han Z, Sagum CA, Takizawa F, et al. Ubiquitin ligase WWP1 interacts with Ebola virus VP40 to regulate Egress. J Virol 2017; 91. doi: 10.1128/JVI.00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vale-Costa S, Amorim MJ. Recycling endosomes and viral infection. Viruses 2016; 8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol 2004; 5:121–32. [DOI] [PubMed] [Google Scholar]
- 22. Gromov PS, Celis JE, Hansen C, Tommerup N, Gromova I, Madsen P. Human rab11a: transcription, chromosome mapping and effect on the expression levels of host GTP-binding proteins. FEBS Lett 1998; 429:359–64. [DOI] [PubMed] [Google Scholar]
- 23. Amorim MJ, Bruce EA, Read EK, et al. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J Virol 2011; 85:4143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Avilov SV, Moisy D, Munier S, Schraidt O, Naffakh N, Cusack S. Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. J Virol 2012; 86:1433–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J Virol 2011; 85:6117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Momose F, Sekimoto T, Ohkura T, et al. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS One 2011; 6:e21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chambers R, Takimoto T. Trafficking of Sendai virus nucleocapsids is mediated by intracellular vesicles. PLoS One 2010; 5:e10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stone R, Hayashi T, Bajimaya S, Hodges E, Takimoto T. Critical role of Rab11a-mediated recycling endosomes in the assembly of type I parainfluenza viruses. Virology 2016; 487:11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hollinshead M, Johns HL, Sayers CL, Gonzalez-Lopez C, Smith GL, Elliott G. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J 2012; 31:4204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Diederich S, Sauerhering L, Weis M, et al. Activation of the Nipah virus fusion protein in MDCK cells is mediated by cathepsin B within the endosome-recycling compartment. J Virol 2012; 86:3736–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varthakavi V, Smith RM, Martin KL, et al. The pericentriolar recycling endosome plays a key role in Vpu-mediated enhancement of HIV-1 particle release. Traffic 2006; 7:298–307. [DOI] [PubMed] [Google Scholar]
- 32. Brock SC, Goldenring JR, Crowe JE Jr. Apical recycling systems regulate directional budding of respiratory syncytial virus from polarized epithelial cells. Proc Natl Acad Sci U S A 2003; 100:15143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Utley TJ, Ducharme NA, Varthakavi V, et al. Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11-FIP2. Proc Natl Acad Sci U S A 2008; 105:10209–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bruce EA, Digard P, Stuart AD. The Rab11 pathway is required for influenza A virus budding and filament formation. J Virol 2010; 84:5848–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pereira LE, Clark J, Grznarova P, et al. Direct evidence for intracellular anterograde co-transport of M-PMV Gag and Env on microtubules. Virology 2014; 449:109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sfakianos JN, LaCasse RA, Hunter E. The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic 2003; 4:660–70. [DOI] [PubMed] [Google Scholar]
- 37. Arnaud F, Murcia PR, Palmarini M. Mechanisms of late restriction induced by an endogenous retrovirus. J Virol 2007; 81:11441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog 2012; 8:e1002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spurgers KB, Alefantis T, Peyser BD, et al. Identification of essential filovirion-associated host factors by serial proteomic analysis and RNAi screen. Mol Cell Proteomics 2010; 9:2690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen W, Feng Y, Chen D, Wandinger-Ness A. Rab11 is required for trans-golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol Biol Cell 1998; 9:3241–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nanbo A, Imai M, Watanabe S, et al. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog 2010; 6:e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nanbo A, Maruyama J, Imai M, et al. Ebola virus requires a host scramblase for externalization of phosphatidylserine on the surface of viral particles. PLoS Pathog 2018; 14:e1006848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nanbo A, Kachi K, Yoshiyama H, Ohba Y. Epstein-Barr virus exploits host endocytic machinery for cell-to-cell viral transmission rather than a virological synapse. J Gen Virol 2016; 97:2989–3006. [DOI] [PubMed] [Google Scholar]
- 44. Nanbo A, Watanabe S, Halfmann P, Kawaoka Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci Rep 2013; 3:1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Watanabe S, Watanabe T, Noda T, et al. Production of novel ebola virus-like particles from cDNAs: an alternative to ebola virus generation by reverse genetics. J Virol 2004; 78:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamayoshi S, Kawaoka Y. Mapping of a region of Ebola virus VP40 that is important in the production of virus-like particles. J Infect Dis 2007; 196(Suppl 2):S291–5. [DOI] [PubMed] [Google Scholar]
- 47. Silvis MR, Bertrand CA, Ameen N, et al. Rab11b regulates the apical recycling of the cystic fibrosis transmembrane conductance regulator in polarized intestinal epithelial cells. Mol Biol Cell 2009; 20:2337–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dozynkiewicz MA, Jamieson NB, Macpherson I, et al. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev Cell 2012; 22:131–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Horgan CP, McCaffrey MW. The dynamic Rab11-FIPs. Biochem Soc Trans 2009; 37:1032–6. [DOI] [PubMed] [Google Scholar]
- 50. Desnos C, Huet S, Darchen F. ‘Should I stay or should I go?’: myosin V function in organelle trafficking. Biol Cell 2007; 99:411–23. [DOI] [PubMed] [Google Scholar]
- 51. Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem 2003; 278:41573–6. [DOI] [PubMed] [Google Scholar]