Abstract
In the budding yeast Saccharomyces cerevisiae, transcription factors (TFs) regulate the periodic expression of many genes during the cell cycle, including gene products required for progression through cell-cycle events. Experimental evidence coupled with quantitative models suggests that a network of interconnected TFs is capable of regulating periodic genes over the cell cycle. Importantly, these dynamical models were built on transcriptomics data and assumed that TF protein levels and activity are directly correlated with mRNA abundance. To ask whether TF transcripts match protein expression levels as cells progress through the cell cycle, we applied a multiplexed targeted mass spectrometry approach (parallel reaction monitoring) to synchronized populations of cells. We found that protein expression of many TFs and cell-cycle regulators closely followed their respective mRNA transcript dynamics in cycling wild-type cells. Discordant mRNA/protein expression dynamics was also observed for a subset of cell-cycle TFs and for proteins targeted for degradation by E3 ubiquitin ligase complexes such as SCF (Skp1/Cul1/F-box) and APC/C (anaphase-promoting complex/cyclosome). We further profiled mutant cells lacking B-type cyclin/CDK activity (clb1-6) where oscillations in ubiquitin ligase activity, cyclin/CDKs, and cell-cycle progression are halted. We found that a number of proteins were no longer periodically degraded in clb1-6 mutants compared with wild type, highlighting the importance of posttranscriptional regulation. Finally, the TF complexes responsible for activating G1/S transcription (SBF and MBF) were more constitutively expressed at the protein level than at periodic mRNA expression levels in both wild-type and mutant cells. This comprehensive investigation of cell-cycle regulators reveals that multiple layers of regulation (transcription, protein stability, and proteasome targeting) affect protein expression dynamics during the cell cycle.
INTRODUCTION
The eukaryotic cell cycle is a complex biological process, in which many regulatory proteins have been characterized. The cell cycle is very well understood in the budding yeast model system Saccharomyces cerevisiae (Hartwell et al., 1974; Elliott and McLaughlin, 1978; Lord and Wheals, 1981). Cyclins, cyclin-dependent kinases (CDKs), and protein degradation machinery trigger and order cell cycle events, and transcription factors (TFs) regulate the abundance of these cell-cycle regulators by activating “just-in-time” phase-specific gene expression (Spellman et al., 1998; Simon et al., 2001; Lee et al., 2002; Pramila et al., 2006; Orlando et al., 2008, Cho et al., 2017a; reviewed in Haase and Wittenberg, 2014). Cell-cycle machinery is highly conserved across eukaryotes (Lee and Nurse, 1987; Elledge and Spottswood, 1991; Ninomiya-Tsuji et al., 1991), and programs of periodic genes have been identified in many species (Ishida et al., 2001; Rustici et al., 2004; Menges et al., 2005; Grant et al., 2013, Kelliher et al., 2016). Despite the observation that many regulatory components of the eukaryotic cell cycle are periodically transcribed, it is largely unknown whether periodic mRNAs are also periodically expressed at the protein level.
Posttranscriptional modifications such as phosphorylation and ubiquitination are known to affect protein activity and rate of turnover, respectively, for cell-cycle regulators (Ubersax et al., 2003; Breitkreutz et al., 2010; Swaney et al., 2013). Indeed, several studies have shown that mRNA and protein expression levels can be poorly correlated (Futcher et al., 1999; Gygi et al., 1999; Serikawa et al., 2003; Washburn et al., 2003; Jensen et al., 2006; Lackner et al., 2007; de Godoy et al., 2008; Marguerat et al., 2012; Li et al., 2014; reviewed in Vogel and Marcotte, 2012). However, most of these studies have analyzed the relationship between mRNA and protein abundance from steady state populations of cells, without considering the timing or kinetics of mRNA and protein turnover in the cell cycle. Only a few previous studies have profiled proteome dynamics for S. cerevisiae, Schizosaccharomyces pombe, and human cells at time points corresponding to the major cell-cycle phases (Flory et al., 2006; Carpy et al., 2014; Ly et al., 2014). Gene expression dynamics have been investigated with denser sampling over time for a small number of cell-cycle proteins by our group and others, which demonstrated that S. cerevisiae cell-cycle protein expression closely followed periodic mRNA expression levels (Ball et al., 2011).
While microarray and RNA-sequencing technologies enable complete quantitation of the S. cerevisiae transcriptome, a complementary methodology is still lacking for quantitation of the total proteome. ORF-tagging strategies have enabled single-cell fluorescence or immunoblotting analysis of protein expression for individual proteins (Ghaemmaghami et al., 2003; Huh et al., 2003; Kulak et al., 2014; Chong et al., 2015; Yofe et al., 2016), but available tools are not amenable to multiplexing across cell-cycle time. Likewise, unbiased proteomics using liquid chromatography tandem mass spectrometry (LC-MS/MS) and isobaric tagging have achieved nearly complete quantitation of the S. cerevisiae proteome (∼5000 proteins) but require extensive fractionation and are limited to multiplexing of ∼10 samples (Paulo et al., 2015). In contrast, targeted proteomics can offer high sensitivity and quantitative precision and the capability of multiplexed protein quantitation across unlimited numbers of samples (Picotti et al., 2009, 2013; Costenoble et al., 2011; Mirzaei et al., 2013; Soste et al., 2014).
Here, we sought to apply the first targeted proteomic approach to quantifying cell-cycle-dependent protein expression across a large set of S. cerevisiae proteins during the cell cycle.
Specifically, we quantified protein abundance from synchronous cells and compared transcriptome with proteome dynamics during the cell cycle. Our study is the most densely sampled proteomics data set across the cell cycle (20 or more time points), enabling us to quantify detailed cell-cycle dynamics from ∼45 TFs and regulatory proteins in S. cerevisiae. We found that many proteins match their respective mRNA dynamics, with some interesting exceptions. Furthermore, we profiled the transcriptome and proteome from mutant cells and demonstrated that periodic cyclin/CDK and periodic ubiquitin ligase activity are required for the dynamic expression of some cell-cycle proteins.
RESULTS
Targeted mass spectrometry method development
We developed a targeted proteomic assay for multiplexed quantitation of 45 cell-cycle proteins and four constitutively expressed controls for cytoplasmic (Rim11, Vps9) and nuclear (Cic1, Taf12) localization (Supplemental Table 1). To select peptides for quantitation, we used available discovery-based and targeted proteomic data from PeptideAtlas and other sources, along with previously published guidelines for optimal peptide selection (see Materials and Methods; Desiere et al., 2006; Mirzaei et al., 2013). In the absence of prior experimental data, we chose peptide sequences that were selected by Aebersold and colleagues to quantify the entire yeast proteome (Picotti et al., 2013). In total, 149 stable isotope-labeled (SIL) peptides were synthesized to be used as internal standards for identification and quantitation of the 49 targets (Supplemental Table 1).
To test the assay, SIL peptides were spiked into whole-cell tryptic digests of asynchronous wild-type S. cerevisiae cells, and 1 µg of digests was analyzed by LC-MS/MS using parallel reaction monitoring (PRM), a highly sensitive targeted proteomic approach. Native yeast peptides were identified based on the retention time and MS/MS spectra of the SIL peptide standards. After removing targets that had poor reproducibility across triplicate analyses or were undetectable above noise, we were able to quantify 38 peptides belonging to 22 proteins (only 45% of the proteins of interest; see Supplemental File 1). Because many cell-cycle regulators are transiently expressed in specific phases of the cell cycle, we hypothesized that undetectable proteins in asynchronous yeast samples were diluted below the limits of detection.
Many cell-cycle regulators exhibit dynamic protein expression during a wild-type cell cycle
We previously profiled transcriptome dynamics from wild-type budding yeast cells across multiple cell cycles using RNA sequencing, sampling every 5 min (Kelliher et al., 2016). We applied our MS method to cells grown under identical conditions to compare cell cycle-dependent protein expression (Materials and Methods). Cells were collected over time to monitor the budding index (a marker for cell cycle entry) and to extract protein. Protein expression was quantified by PRM in two replicate time series (sampling every 7 min for 20 total time points). For each of the two biological replicate experiments, QC samples were designed by pooling equal amounts of the time points for internal technical replicates. We targeted two peptides per protein of interest to maximize MS duty cycle and sensitivity, based on measurements from method development and the pooled QC samples. For each biological replicate, the time series samples were analyzed in a random order and were interspersed with five analyses of the QC pools.
Using the SIL peptides as internal references, we selected interference-free product ions for quantitation of each of the peptides (Supplemental Table 1) and normalized the expression of native proteins to the SIL peptide standards (Materials and Methods). In total, we quantified ∼80 peptides corresponding to 48 proteins across each of the experiments. A greater number of peptides were identified and quantified in the time-series samples than in asynchronous cells, likely due to high, but transient expression of target proteins at specific points in the cell cycle (Supplemental File 1). We further filtered the data to include only peptides that were reproducibly quantified across QC pools and/or between the two biological replicate experiments. In addition, we analyzed expression-level noise between peptides using the QC pools and individual samples over the two biological replicates (Supplemental Table 2). On the basis of these multiple factors, we assigned the highest confidence to the quantitation of 44 peptides belonging to 31 unique proteins (excluding the four nuclear and cytoplasmic controls, which were detected with high confidence but were not dynamically expressed during the cell cycle).
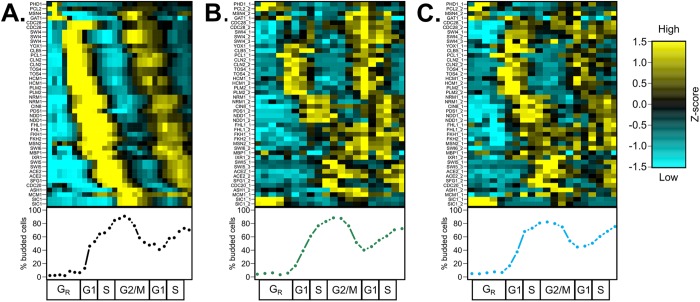
To compare transcriptome and proteome dynamics, we aligned the time-series data to a common cell-cycle timeline using the CLOCCS algorithm (Orlando et al., 2007; Supplemental File 1), and the relative expression of mRNA and protein was visualized across the 31 cell-cycle genes. Qualitatively, many proteins appear to follow their respective RNA dynamics with some delay (Figure 1), indicating that the dynamic transcriptome is generally a good proxy for protein dynamics. Our results support previous immunoblotting experiments for 14 individual cell-cycle proteins (Supplemental Table 1) and demonstrate cell-cycle dynamics for 17 proteins for the first time.
FIGURE 1:
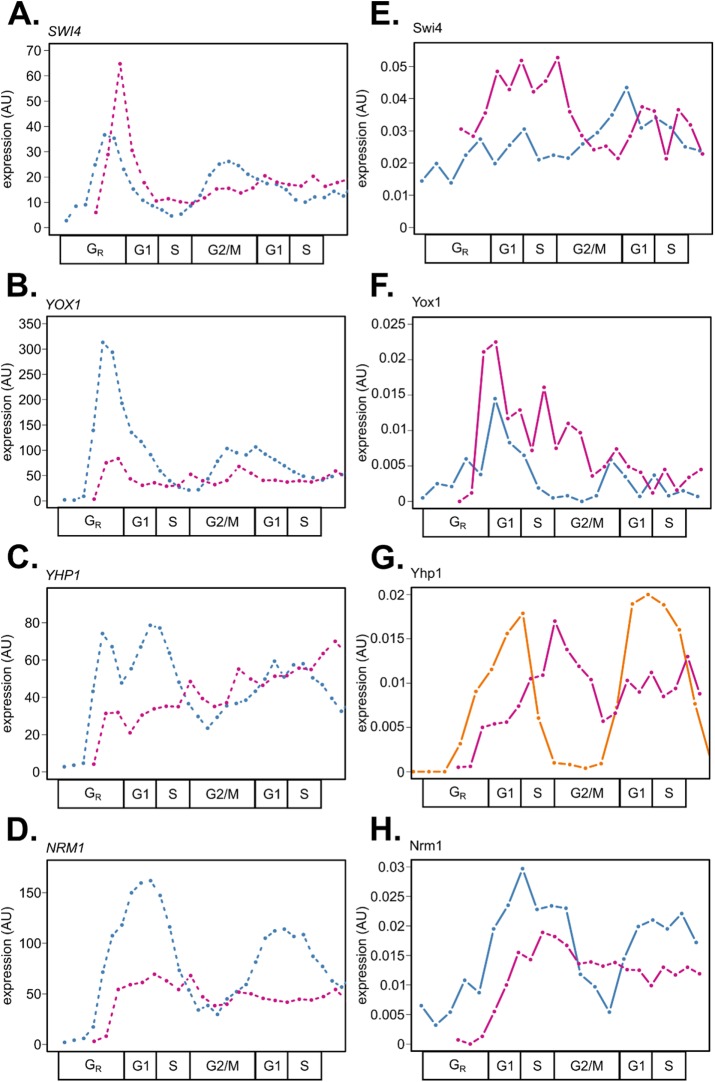
Proteome dynamics of cell-cycle regulators follow the transcriptome with delay during the cell cycle in S. cerevisiae. Wild-type budding yeast cells were grown in 2% YEPD rich media, synchronized by alpha-factor mating pheromone, released into YEPD, and monitored over ∼2 cell cycles. Samples were collected every 5 min for RNA sequencing (Kelliher et al., 2016) (A) or every 7 min for total protein extraction (B, C). Population synchrony was monitored by counting at least 200 cells per time point for the presence or absence of a bud (bottom). A total of 31 high-confidence proteins (44 total peptides) are shown, with multiple high-confidence peptides per protein for Cdc28, Swi4, Cln2, Tos4, Hcm1, Plm2, Nrm1, Ndd1, Fhl1, Swi5, Ace2, and Sic1. Genes and proteins were ordered on the y-axis by peak time of mRNA expression (A). Transcript and protein levels are depicted as z-score changes relative to expression mean in the respective data sets, where values represent the number of standard deviations away from the mean. Each column represents a lifeline point on a common cell-cycle timeline determined by the CLOCCS algorithm (Supplemental File 1). Individual line plots from Figure 1 are shown for core cell-cycle TFs (Supplemental Figure 1), non-strongly-cycling regulators (Supplemental Figure 2), and periodic regulators (Supplemental Figure 3). Eleven regulators (Gat1, Cdc28, Swi4, Fhl1, Fkh1, Fkh2, Msn2, Swi6, Mbp1, Ixr1, and Mcm1) were somewhat constitutively expressed, and 20 regulators (Phd1, Pcl2, Msn4, Yox1, Clb5, Pcl1, Cln2, Tos4, Hcm1, Plm2, Nrm1, Cin8, Pds1, Ndd1, Swi5, Ace2, Sfg1, Cdc20, Ash1, and Sic1) appeared to be cycling.
Prior to cell-cycle Start, early G1 genes are controlled by the constitutively expressed coactivator Mcm1 and the cycling corepressors Yhp1 and Yox1 (Pramila et al., 2002; Supplemental Figure 1A). At cell-cycle Start, ∼ 200 periodic genes are activated by the heterodimeric TFs SBF (Swi4/Swi6) and MBF (Mbp1/Swi6) (Iyer et al., 2001; Eser et al., 2011; Supplemental Figure 1B) after removal of the early G1 corepressors Whi5 and Stb1 (non-high-confidence proteins; Supplemental Table 2). S-phase genes are then activated by Hcm1 and regulated by the paralogous TFs Plm2 and Tos4 (Horak et al., 2002; Pramila et al., 2006; Supplemental Figure 1C). G2/M genes are controlled by the Swi Five Factor (SFF) complex of TFs: Mcm1, Fkh1, Fkh2, and the cycling co-activator Ndd1 (Reynolds et al., 2003; Supplemental Figure 1D). M/G1 genes are activated by the periodic paralogues Ace2 and Swi5 (Di Talia et al., 2009; Supplemental Figure 1E). Intriguingly, several of these core cell-cycle TFs did not appear to cycle strongly in abundance at the protein level in comparison with the dynamic behavior of the corresponding RNA expression. An additional subset of cell-cycle regulators were not periodically expressed by visual inspection (Supplemental Figure 2, A–F), totaling over 10 proteins that did not appear to match cycling transcript dynamics (Figure 1). A majority of cell-cycle regulators (20/31, 64.5%) did appear to cycle at the protein level (Figure 1; Supplemental Figure 3).
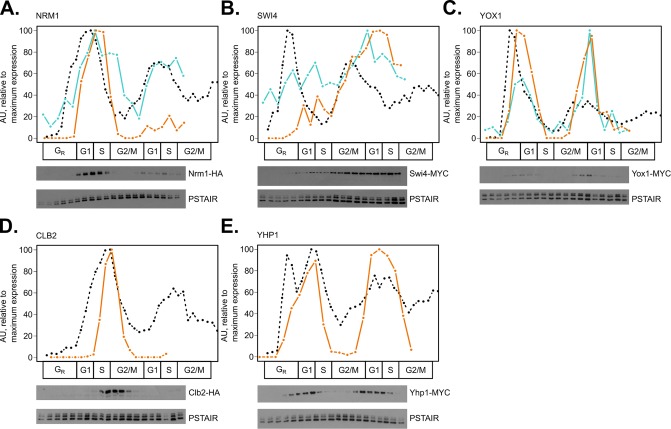
Specifically, the core cell-cycle TFs Swi4, Mbp1, Swi6, Fkh1, and Fkh2 did not appear strongly periodic at the protein level (Supplemental Figure 1, B and D). Fkh1-2 function in the SFF complex, whose coactivator Ndd1 is periodic at the protein level. Thus, the periodic activity of the SFF complex is gated by Ndd1 expression (Reynolds et al., 2003). On the other hand, all subunits of the SBF and MBF TF activator complexes did not appear to be strongly periodically expressed at the protein level despite periodic mRNA expression (Orlando et al., 2008; Kelliher et al., 2016). MBF is inactivated during the S-phase by its corepressor Nrm1 (de Bruin et al., 2006). The SWI4 transcript is repressed by the paralogous TFs Yhp1 and Yox1 (Pramila et al., 2002), and the SBF complex is inactivated and removed from target gene promoters later by B-type cyclin/CDK phosphorylation (Amon et al., 1993). Two key negative regulators of SBF and MBF, Clb2 and Yhp1, were not detected with high confidence in the wild-type PRM assay. Time series immunoblotting experiments in triplicate suggest that the proteins are periodic and match the dynamics of their cognate mRNAs (Figure 2, D and E; Supplemental Figure 4, D and E). Additionally, we validated the protein expression dynamics of Swi4, Nrm1, and Yox1 TFs by immunoblotting (Figure 2, A–C; Supplemental Figure 4, A–C). While Nrm1 and Yox1 appeared to match the dynamics of their respective mRNAs, Swi4 did not appear strongly periodic. Taking these together, it appeared that repressor protein dynamics more closely matched mRNA dynamics than activator TFs at G1/S phase.
FIGURE 2:
Method validation and supplementation of targeted mass spectrometry in comparison with time series immunoblots. Line plots for mRNA expression (black, dashed), representative Western blot protein expression (orange, solid), and representative PRM peptide expression (blue, solid) were aligned on a common cell-cycle timeline using CLOCCS and plotted. Wild-type cells expressing Nrm1-HA3 (A), Swi4-13MYC (B), Yox1-13MYC (C), Clb2-HA (D), or Yhp1-13MYC (E) were grown in 2% YEPD media, synchronized by alpha-factor mating pheromone, released into YEPD, and monitored over ∼2 cell cycles. Samples were collected every 7 min for total protein extraction. Protein immunoblots were normalized to Cdc28/Pho85 (PSTAIR; constitutive levels over the cell cycle) with ImageJ. One representative Western blot is shown for each triplicate set of experiments (orange lines). To assess reproducibility between PRM and immunoblotting, Western blot data were compared with targeted mass spectrometry peptide data (blue lines) for NRM1_1 from PRM replicate 2, A; SWI4_1 from PRM replicate 1, B; and YOX1_1 from PRM replicate 1, C. Transcript expression, peptide light/heavy ratios, and Western blot data were scaled to maximum expression for each gene or protein ([0, 100] linear scale).
We next sought a method for measuring similarity of mRNA and protein expression curves to quantify the extent to which the dynamics of a transcript matches the dynamics of its protein and to estimate the temporal delay between mRNA and protein (i.e., the degree and nature of posttranscriptional regulation on cell-cycle proteins). Inspired by the algorithm JTK_CYCLE, which was designed to identify rhythmic dynamics in time series experiments (Hughes et al., 2010), we adapted the Kendall tau distance function on ranked lists to the problem of measuring curve-shape similarity (Materials and Methods). The Temporal Alignment Kendall-Tau (TAKT) algorithm produces measures of curve-shape similarity and a significance score to assess the degree of dynamic similarity between mRNA–protein pairs over cell-cycle time. Using the TAKT method, we found that 81 out of 94 (86.2%) total peptides or immunoblot measurements (44 high-confidence peptides in two biological replicates, two proteins quantified by Western blot in three biological replicates) were well correlated with mRNA expression (Supplemental Table 3; empirical p value < 0.05). Thus, a majority of protein time series curves had a better TAKT similarity score to their cognate mRNA curves than was achieved by at least 95% of randomized mRNA expression profiles (Materials and Methods).
In some cases, only a subset of the peptides and/or biological replicates associated with a given protein exhibited strong dynamic similarity to the corresponding mRNA dynamics. Eleven discordant RNA–protein pairs were identified by eye (Figure 1: Cdc28, Fhl1, Fkh1, Fkh2, Gat1, Ixr1, Mbp1, Mcm1, Msn2, Swi4, and Swi6), and all had at least one peptide and/or biological replicate with a TAKT score of p > 0.01, indicating that these proteins may be regulated posttranscriptionally (Supplemental Figures 1 and 2). That said, only Cdc28 (both peptides) and Msn2 (one peptide) were significantly dynamically different from mRNA expression in all measured peptides across biological replicates (Supplemental Table 3). Seven proteins (Fkh1, Fkh2, Gat1, Ixr1, Mbp1, Mcm1, and Swi6) were quantified with one peptide, and only one biological replicate of that peptide suggested discordant RNA–peptide expression. Two proteins (Fhl1, Swi4) had 2–3 high-confidence peptides and only one representative peptide with a discordant TAKT score (FHL1_1 in replicate 1; SWI4_3 in replicate 2; Supplemental Figure 5). Thus, these 11 cell-cycle proteins display some variability in the degree of correlation between periodic mRNA expression and protein abundance (Orlando et al., 2008; Kelliher et al., 2016) and are not clearly cycling at the protein level (Supplemental Figures 1 and 2; Supplemental Table 3).
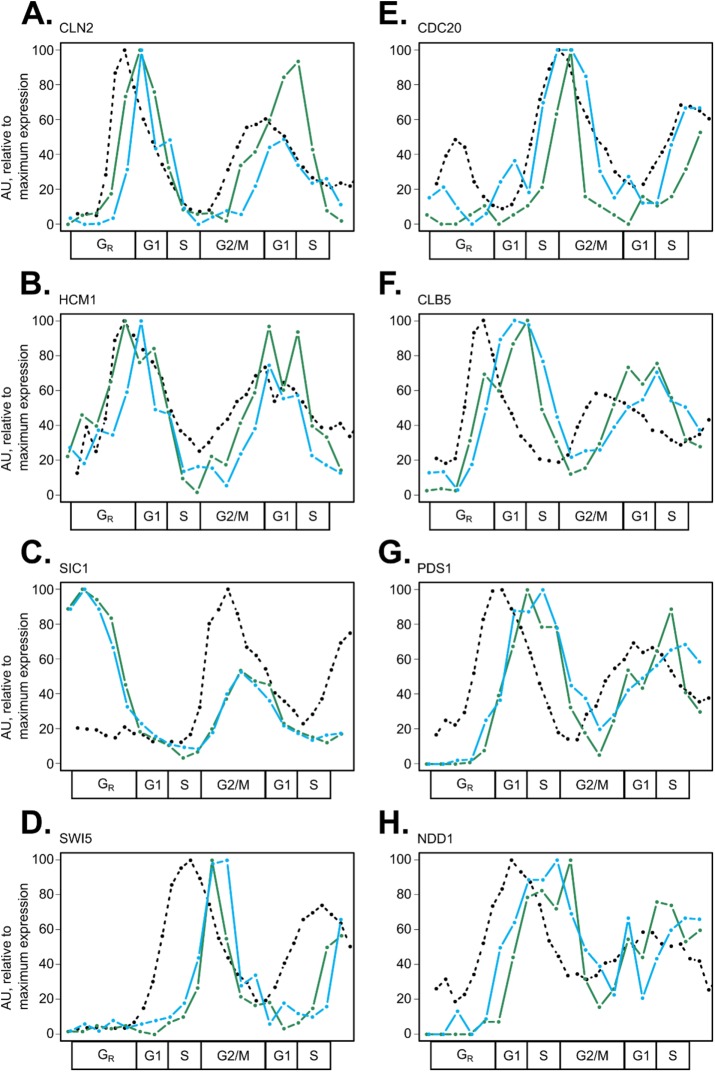
A majority of cell-cycle proteins did appear to be strongly influenced by periodic mRNA expression, with varying amounts of temporal shift (Figure 3). Thus, we mined the literature for previously reported and predicted protein degradation mechanisms of cell-cycle regulators of interest (Supplemental Table 4), and we quantified the delay between mRNA and protein expression using the TAKT algorithm (Supplemental Table 5; Materials and Methods). This analysis revealed that there was not one consistent lag time between mRNA and protein synthesis, supporting the conclusion that specific periodic proteins may be subject to posttranscriptional and/or posttranslational regulation. We observed that proteins targeted for destruction at specific times during the cell cycle, such as Clb5 and Pds1, had long delay times from mRNA to protein, yet exhibited curve shape dynamics very similar to that of their cognate mRNA in cycling wild-type cells (Figure 3, F and G; Supplemental Table 3).
FIGURE 3:
Cell-cycle proteins targeted for destruction by E3 ubiquitin ligases have unique expression features in wild-type cells. RNA expression and peptide light/heavy ratios were scaled to the maximum value for each gene or protein ([0, 100] linear scale). Line plots for mRNA expression (black, dashed) and biological replicates of wild-type peptide expression (replicate 1 in green, replicate 2 in blue) are shown for canonical SCF targets: CLN2_1 (A), HCM1_1 (B), SIC1_1 (C), and SWI5_3 (D) or APC/C targets: CDC20_1 (E), CLB5_1 (F), PDS1_2 (G), and NDD1_1 (H). When multiple peptides per protein were detected (Figure 1), the peptide with lowest noise levels was selected (Supplemental Table 2).
To further investigate the effect of targeted protein destruction on protein expression dynamics, we visualized SCF targets (Cln2, Hcm1, Sic1, and Swi5; Figure 3, A–D), APC/C targets (Cdc20, Clb5, and Pds1; Figure 3, E–G), and targets of both (Ndd1; Figure 3H) over the cell cycle (Supplemental Table 4). SCF-targeted proteins are turned over at various times throughout the cell cycle, and this process is regulated by target phosphorylation (Figure 3, A–D). APC/C targets were turned over at a precise time during the G2/M phase, and this destruction timing lined up with peak expression of the APC/C cofactor, Cdc20 (Figure 3E). Interestingly, the accumulation of some APC/C target proteins also appeared to be regulated, with a substantial delay between increases in mRNA expression and subsequent protein expression (Figure 3, G and H). This result is supported by previous work on the role of APC/C-Cdh1 in inhibiting the accumulation of select G1/S cell-cycle proteins, thus enforcing the timing of S-phase entry (Huang et al., 2001; Yeong et al., 2001; Yuan et al., 2014). SCF targets had a shorter average mRNA-to-protein delay time of ∼8.1 ± 4.8 min (assuming a 70-min cell cycle, 11.6 ± 6.9 cell-cycle timeline points) compared with ∼13.1 ± 11.3 min (18.7 ± 16.1 cell-cycle points) for APC/C targets (Supplemental Table 5).
In the absence of periodic cyclin/CDK and ubiquitin ligase activities, some cell-cycle proteins continue to be dynamically expressed
To query the importance of periodic SCF and APC/C activity in regulating cell-cycle protein expression, we profiled clb1-6 cells where these E3 ubiquitin ligase complexes should not have periodic activity, but many cell-cycle genes continue to be periodically transcribed (Haase and Reed, 1999; Orlando et al., 2008). Upon synchronization and induction, cells lacking B-type cyclins become physically arrested at the G1/S phase border and cannot initiate DNA replication (Haase and Reed, 1999). Cell-cycle proteins that are phosphorylated by B-cyclin/CDKs and targeted for degradation by SCF should therefore be stabilized. APC/C-Cdh1 should be active only at the beginning of the time series (as observed in wild-type cells) and subsequently inactivated by Cln/CDKs and other kinases (Zachariae et al., 1998; Hall et al., 2004). Later in time, APC/C-Cdc20 should not become activated because functional Cdc20 requires B-cyclin/CDK phosphorylation (Rudner and Murray, 2000; Rahal and Amon, 2008). Therefore, clb1-6 mutant protein expression dynamics should be largely dependent on mRNA dynamics and protein half-life.
GAL-CLB1 clb1-6 cells were cultured in YEPG media, arrested in G1 phase using alpha-factor mating pheromone, supplemented with dextrose to inhibit CLB1 expression, and then released into YEPD media at 30°C. Cells were collected over time to monitor the rebudding index, isolate mRNA, or extract protein (Materials and Methods). Replicate time series experiments were aligned to a common cell-cycle timeline using the CLOCCS algorithm (Orlando et al., 2007; Supplemental File 1).
Cell-cycle regulatory proteins were less correlated with their respective mRNA precursors in clb1-6 mutant cells by TAKT score than wild type, with only 13 positively correlated RNA–peptide pairs in both biological replicates, representing 11 unique proteins (Supplemental Table 3 and Supplemental Figure 6). This included a subset of core cell-cycle TFs (Swi4, Swi6, Nrm1, Ndd1, Ace2, and Swi5), which were positively correlated with mRNA expression in mutant cells (Supplemental Figure 6, B, D, and E). Poorer RNA–protein correlation scores in mutant compared with wild-type cells were likely not due to noise in protein expression data, as the magnitude of noise values was similar between experiments (Supplemental Table 2).
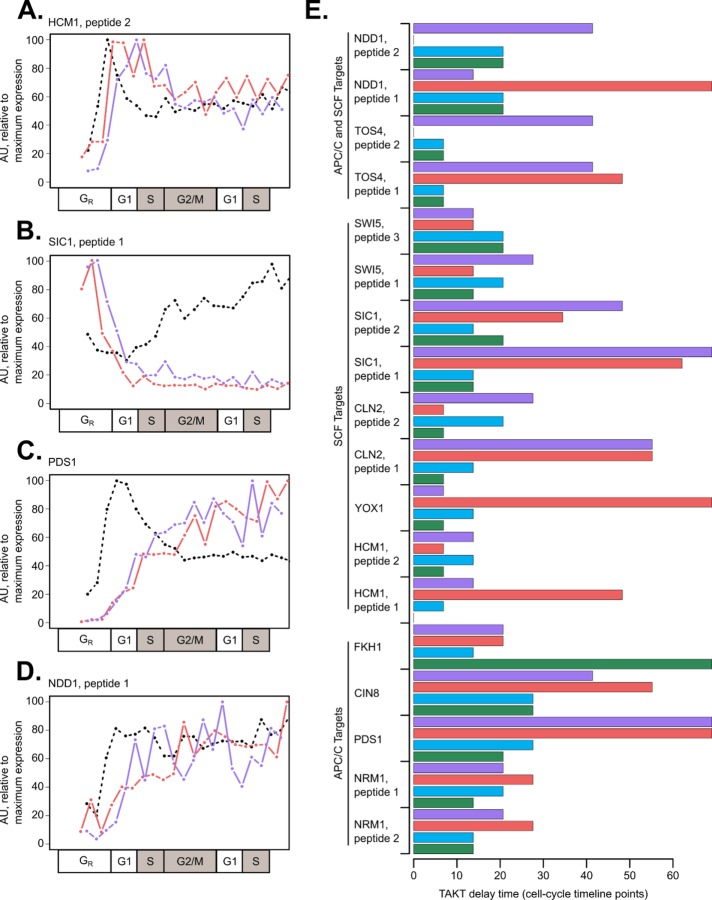
We hypothesized that lack of periodic protein destruction could explain the decreased correlation for some RNA–protein pairs in mutant cells as compared with wild type. In support of this hypothesis, both APC/C and SCF targets had variable degradation kinetics in mutant cells as compared with wild type (Figure 4, A–D). We posited that SCF targets requiring Clb/CDK phosphorylation would be stabilized and that SCF targets requiring Cln/CDK phosphorylation would be highly unstable in mutant cells. Consistent with this expectation, Sic1 exhibited only one early peak of protein expression in clb1-6 cells, which suggested persistent Cln/CDK phosphorylation and SCF degradation later in the time series (Figure 4B). On the other hand, Hcm1 was not turned over to low levels in clb1-6 cells (Figure 4A), which is consistent with the stabilization of Hcm1 mutants lacking B-type cyclin/CDK phosphorylation sites (Landry et al., 2014). Finally, canonical APC/C target proteins accumulated after APC/C-Cdh1 removal and were not destroyed later, which is likely because APC/C-Cdc20 is inactive in clb1-6 mutants (Figure 4, C and D). The absolute protein expression levels of these APC/C targets were also increased in clb1-6 compared with wild type, where Ndd1 and Pds1 peptide abundance increased more than fourfold (0.0374/0.00682 and 0.0306/0.00653, respectively, for Ndd1 and Pds1; average light/heavy ratios from Supplemental Tables 6 and 7). Delays between mRNA and protein expression were generally greater in mutant cells than in wild type (Figure 4E; Supplemental Table 5).
FIGURE 4:
E3 ubiquitin ligase targets display altered dynamics and RNA-to-protein delay times in clb1-6 mutant cells. RNA expression and peptide light/heavy ratios were scaled to the maximum value for each gene or protein ([0, 100] linear scale). Line plots for mRNA expression (black, dashed) and biological replicates of clb1-6 mutant peptide expression (replicate 1 in red, replicate 2 in purple) are shown for canonical SCF targets: HCM1_2 (A), SIC1_1 (B) or APC/C targets: PDS1_2 (C), and NDD1_1 (D). When multiple peptides per protein were detected, the peptide with lower noise levels was selected (Supplemental Table 2). In the cell-cycle timeline for clb1-6 cells, S and G2/M phases are shown as gray boxes to indicate that B-cyclin mutant cells are physically arrested at the G1/S border. RNA-to-peptide delay times were calculated using the TAKT algorithm and shown in cell-cycle timeline points (wild-type replicates: green and blue bars; clb1-6 replicates: red and purple bars) (E). RNA–protein pairs are grouped based on putative targeted degradation mechanism (Supplemental Table 4). The average time delay for peptides shown in clb1-6 mutant cells was 33.7 ± 22.0 cell-cycle points, compared with 16.5 ± 11.3 cell-cycle points in wild-type cells (E).
Repressor TFs are more periodically expressed at the protein level than G1/S activator TFs in both wild-type and mutant cells
After commitment to the cell cycle, ∼200 genes are periodically activated at G1/S phase by the TF complexes SBF and MBF in S. cerevisiae (Iyer et al., 2001; Eser et al., 2011). These TFs are functionally analogous to the E2F family of TFs in mammalian cells (reviewed in Bertoli et al., 2013). The subunits of SBF and MBF (Swi4, Swi6, and Mbp1) are periodically transcribed during the cell cycle (Spellman et al., 1998). The program of G1/S genes is periodically activated in both wild-type and clb1-6 mutant cells, suggesting that the activity of SBF and MBF is periodic under both conditions (Orlando et al., 2008).
We compared the mRNA and protein expression dynamics of G1/S activator (Swi4) and repressor (Yox1, Yhp1, Nrm1) TFs in wild-type and clb1-6 mutant cells (Figure 5). As observed previously for wild-type cells (Figures 1 and 2), repressor TFs were more completely degraded and dynamically expressed at the protein level in clb1-6 mutant cells (Figure 5, F–H). These results are consistent with previous findings that non-CDK-phosphorylatable alleles of Yhp1 and Yox1 continue to cycle with higher protein expression levels (Landry et al., 2014). Because SBF and MBF subunits do not appear to be periodically expressed at the protein level, we posit that the abundance of inhibiting cofactors plays an important role in regulating the TF activity of SBF and MBF. The SWI4, SWI6, and MBP1 subunits are clearly periodic at the mRNA level (Figure 1; Supplemental Figure 1B; Figure 5A; Orlando et al., 2008; Kelliher et al., 2016), so it is possible that nascent pools of unmodified SBF and MBF proteins are required each cell cycle from periodic mRNA synthesis, while the total protein abundance of SBF and MBF remains constitutive over the cell cycle. Alternatively, the nuclear localization of Swi4, Swi6, and/or Mbp1 could be altered without changing the total protein abundance, which has been demonstrated for Swi6 nuclear export following Clb6/CDK phosphorylation (Geymonat et al., 2004).
FIGURE 5:
Gene expression dynamics of G1/S TFs in wild-type and clb1-6 mutant cells reveal that repressor proteins are more dynamically expressed than activators. RNA and protein expression data sets were aligned on a common cell-cycle timeline using CLOCCS. Line plots for mRNA expression (dashed) and representative peptide expression (solid lines; wild type in blue and clb1-6 mutant in purple) are shown. mRNA expression values (fpkm units) on the y-axes were normalized together for the two data sets (A–D). Peptide expression values (light/heavy ratios) on the y-axes are comparable between the time series data sets because a constant amount of SIL peptides was used in all experiments (E–H). Representative peptides from PRM experiments are SWI4_1 from wild-type replicate 1 and clb1-6 replicate 2, E; YOX1_1 from wild-type replicate 2 and clb1-6 replicate 2, F; YHP1_5 from clb1-6 replicate 2, G; and NRM1_1 from wild-type replicate 2 and clb1-6 replicate 2, H. Yhp1-13MYC protein expression levels in wild-type cells were taken from Figure 2 (G, orange line). The immunoblotting data were scaled relative to the maximum value for the experiment ([0, 100] linear scale) followed by scaling to the maximum value to match the PRM data from clb1-6 cells ([0, 0.02] linear scale, arbitrary units; G).
DISCUSSION
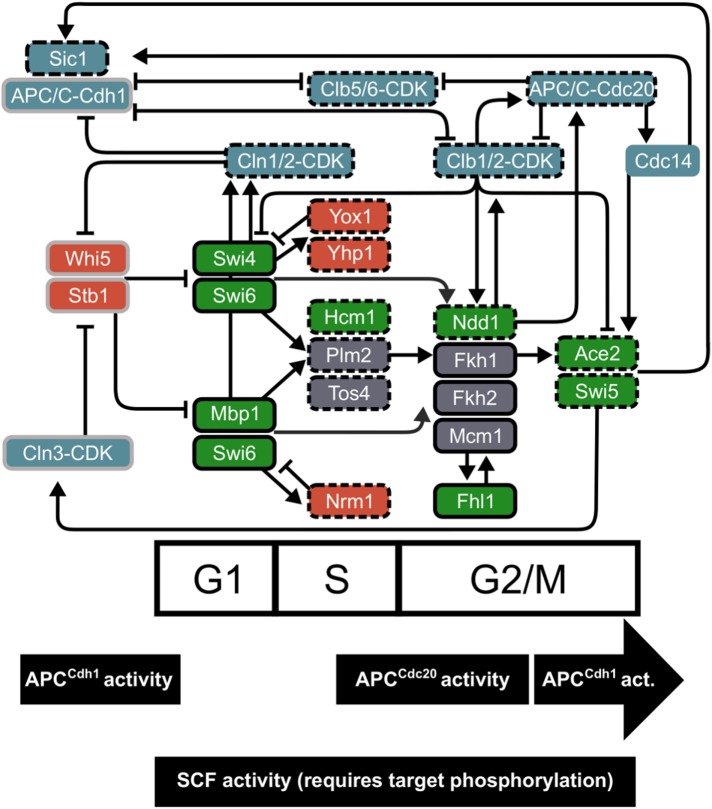
During the cell cycle, an interconnected network of transcription factors, cyclin/CDK complexes, and E3 ubiquitin ligase machinery regulates the order of events and periodic gene expression (Figure 6). Here, we have provided further evidence for the importance of post-transcriptional modifications of cell-cycle regulatory proteins (Supplemental Table 4). Many quantitative models have been developed to explain how a network of TFs regulates periodic gene expression during the S. cerevisiae cell cycle (Orlando et al., 2008; Sevim et al., 2010; Simmons Kovacs et al., 2012; Hillenbrand et al., 2016; Cho et al., 2017a). The proposal that cascading transcriptional activation occurs through a TF network relies on the assumption that mRNA expression is a reliable proxy for cell-cycle protein activity. Although not all periodic mRNAs of network regulators produced periodic proteins (Figure 1), at least one component of each TF complex (e.g., SFF) or TF family (e.g., Swi5/Ace2) exhibits periodic dynamics (Figure 6), which still supports the TF network models. The interesting exceptions appear to be the G1/S activator complexes SBF and MBF. However, the negative regulators of these complexes (Nrm1, Yhp1, Yox1, and Clb2) all exhibit protein dynamics that are a strong match to their periodic mRNAs (Figures 1 and 2; Supplemental Figure 1; Supplemental Table 3; Orlando et al., 2008; Kelliher et al., 2016). Consistent with the most current cell-cycle models (Cho et al., 2017b), negative feedback can generate a pulse of transcriptional activation in the absence of a strong drop in abundance of SBF/MBF components.
FIGURE 6:
An integrated cell-cycle network includes windows of targeted E3 ubiquitin ligase activity. Cell-cycle ordering is maintained by cyclin/CDK activity, an interconnected network of transcription factors, and E3 ubiquitin ligase activity. Approximate windows of peak degradation machinery are shown based on wild-type data (Figure 3). Periodic TFs (activators in green, repressors in red) are placed on the cell-cycle timeline approximately by peak mRNA expression. In blue, G1 (Cln) and B-type (Clb) cyclin/CDKs, APC/C, Cdc14, and Sic1 regulate each other and TFs in the network. Edges from TFs represent evidence for transcriptional regulation: ChIP-chip data for TF binding and/or genetic evidence for regulation type (compiled in Orlando et al., 2008; McGoff et al., 2016; Cho et al., 2017a). Edges between regulatory proteins and TFs represent protein-level modifications (e.g., phosphorylation or ubiquitination). Pointed arrows indicate activation, and blunted arrows mark repression or protein degradation. Proteins outlined in black were detected with high confidence in our study (Supplemental Table 2), and gray indicates low-confidence peptides. Dashed outlines mark periodic cell-cycle regulators, and solid boxes represent more stable TF expression. Paralogues from the whole genome duplication include Yhp1 and Yox1, Plm2 and Tos4, Fkh1 and Fkh2, and Ace2 and Swi5. Complexes of TFs include SBF (Swi4 and Swi6), MBF (Mbp1 and Swi6), and SFF (Mcm1, Fkh1-2, and Ndd1).
We also profiled RNA and protein expression from clb1-6 mutant cells and demonstrated that a set of cell-cycle regulators continued to correlate with mRNA expression independent of B-type cyclin/CDK activity (Supplemental Table 3; Supplemental Figure 6), allowing some periodic transcription to persist in clb1-6 cells (Haase and Reed, 1999; Orlando et al., 2008; Cho et al., 2017a). Other cell-cycle regulator proteins depended on periodic posttranscriptional regulatory mechanisms and were not expressed in a way consistent with their mRNA expression pattern in mutant cells (Supplemental Table 3; Figure 4E), highlighting a role for ubiquitin ligases and other posttranscriptional mechanisms in the control of cell cycle network components. Future quantitative cell-cycle modeling work could improve both wild-type and clb1-6 mutant gene regulatory networks by fitting both RNA and protein expression levels from this study. Taken together, our results show that redundant layers of regulation control the expression of cell-cycle proteins during the yeast cell cycle.
Here, we demonstrated that there is not a single delay time for RNA-to-protein synthesis for the cell-cycle regulators of interest. Along with previous work (Ball et al., 2013), this finding has implications for quantitative models where biochemical rate parameters are assumed to be equal across network components. We estimate that the transcription–translation delay time for the average dynamic yeast protein is ∼14 min with a 13-min SD (Supplemental Table 5). Variance in delay times could indicate mechanisms that differentially control translation or protein stability. Proteins targeted for degradation by the SCF complex had short durations of protein expression in comparison with mRNA (Supplemental Table 5), while the protein accumulation of canonical APC/C targets was gated by both Cdh1 and Cdc20 activity (Figure 3, F–H). Some cell-cycle regulators are turned over by multiple mechanisms (Supplemental Table 4), which further indicates redundancy in cell-cycle control. We posit that redundant destruction mechanisms buffer the cell-cycle regulatory network against ectopic expression of TFs or other cell-cycle proteins in the incorrect phase. This redundancy model could be tested by synchronizing yeast cells, constitutively expressing a cell-cycle TF(s), and querying the degree to which downstream target gene expression is altered.
Curiously, we observed that some proteins did not damp in expression level as much as the respective RNA in the second cell cycle (e.g. Figure 3, A and F). After synchronization, populations of budding yeast cells are expected to damp in gene expression levels over time due to asymmetric cell division between mother and daughter cells (Guo et al., 2013). This protein expression “anti-damping” phenomenon is likely not due to data normalization, as immunoblotting results (where proteins of interest were normalized to a constitutive band) also showed evidence for lack of protein expression damping (Figure 2; Supplemental Figure 4). This anti-damping phenomenon could be explained by previous findings that translation may be more efficient in the second cell cycle after release from alpha-factor synchronization (Serikawa et al., 2003; Goranov et al., 2009).
Currently, DNA or RNA genomic technologies are used more frequently than mass spectrometry technologies to assay gene expression. Therefore, we do not have a complete understanding of the noise models and/or all sources of variance in proteomic data sets. We took advantage of both time series sampling and biological replicate experiments to quantify noise in our PRM data sets (Supplemental Table 2). We dealt with the technical noise in the mass spectrometry data by targeting multiple peptides per protein and by quantifying multiple transitions per peptide (Supplemental Table 1). Future experiments will further our understanding of thresholds, sources of bias, and limits of detection in targeted mass spectrometry.
The current model of the budding yeast cell-cycle network is that unstable TF proteins sequentially activate a large program of gene expression each cell cycle. Future work could further interrogate E3 ubiquitin ligase mutants (SCF, APC/C, proteasome inhibitor treatment, etc.) to query the effects of stabilizing cell-cycle proteins and the resulting gene expression dynamics. The abundance of cell-cycle regulators is only part of the complete mechanism—localization, TF complex formation, and affinity for DNA binding sites require further exploration to fully characterize the dynamics and biological parameters of the yeast cell-cycle network.
MATERIALS AND METHODS
Yeast strains, cultures, and synchronization
Saccharomyces cerevisiae strains are derivatives of BF264-15D MATa bar1. Strains were constructed using standard yeast methods (Supplemental File 1). Yeast cultures were grown in standard YEP media (1% yeast extract, 2% peptone, 0.012% adenine, 0.006% uracil, and 2% dextrose or galactose).
For alpha-factor synchronization experiments, yeast cells were cultured for 2 d, grown overnight at 30°C to mid–log phase, and on the morning of day 3, arrested using 30 ng/ml alpha-factor for approximately one cell-cycle duration (wild-type cells: 110–115 min; clb1-6 mutant cells: 150–160 min). Wild-type cultures were maintained in YEPD media throughout arrest–release. Mutant PGAL1-CLB1 clb1-6Δ cells were cultured prior to the experiments and during alpha-factor arrest in YEPG media. With 40–45 min remaining in alpha-factor arrest, clb1-6 cells were treated with 20% dextrose to a final concentration of 2% to inhibit Clb1 expression. Synchronized wild-type and clb1-6 cultures were washed and resuspended in fresh, prewarmed YEPD media at 30°C at a concentration of ∼1 × 107 cells/ml. Aliquots were taken at each time point and assayed for budding index counts, protein extraction, or RNA extraction.
RNA isolation and RNA-sequencing analyses
Wild-type RNA-sequencing time series data collection and analysis were described previously (Kelliher et al., 2016). Raw RNA-sequencing data from wild-type cells can be found at the NCBI Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) under accession number GSE80474. Wild-type data were renormalized to mutant clb1-6 data from this study.
For clb1-6 cells, total RNA was isolated by acid phenol extraction as described previously (Leman et al., 2014). Samples were sent to the Duke University Sequencing Facility for stranded library preparation. mRNA was amplified and barcoded using KAPA stranded mRNA-Seq library preparation kits, and reads were sequenced in accordance with standard Illumina HiSeq protocols. Libraries of 50 base-pair single-end reads were prepared, and 12 samples were multiplexed for sequencing together in each lane. The RNA-sequencing data analysis pipeline has been described in detail previously (Kelliher et al., 2016). RNA-Seq mapping statistics for this study are presented (Supplemental File 1). Normalized RNA-Seq output (fpkm units) was used in the analyses presented. Raw RNA-Seq data for clb1-6 cells have been submitted to the NCBI GEO database under accession number GSE104904.
Protein isolation and Western blotting
Cell samples were collected on filters (Millipore) at each time point and flash frozen in liquid nitrogen. For each set of time series samples, protein extraction was performed in a cold room at 4°C to inhibit yeast proteases and other enzymes. Cell filters were thawed on ice and then washed in 200 μl of cold 1X phosphate-buffered saline with 0.01% sodium azide (NaN3, which further inhibited yeast cellular processes). Cells were pelleted, resuspended in 1 ml of cold 10% trichloroacetic acid (TCA), and incubated on ice for 5 min. Cells were then resuspended in 100 μl of cold 10% TCA, and acid-washed glass beads (Sigma-Aldrich) were added to each tube. Samples were vortexed for 10 min at 4°C. Cell lysates were then transferred to a fresh tube using a gel-loading pipette tip. Glass beads were washed two times with 100 μl of cold 10% TCA, and cell lysates were transferred to the respective tubes. Lysates were cleared by centrifugation at maximum speed for 10 min at 4°C. After the supernatant was aspirated, protein pellets were washed gently with 1 ml of cold 100% acetone. Aspirated pellets were flash frozen in liquid nitrogen and stored at –80°C.
Protein pellets for Western blot experiments were resuspended in 100 μl of Thorner buffer (8 M urea, 5% SDS, 40 mM Tris-HCl, pH 6.8, 0.1 mM EDTA, 0.4 mg/ml Bromophenol Blue, 1% β-mercaptoethanol) plus 3 μl of 2M Tris Base (unbuffered), boiled for 5 min, and stored at –20°C. SDS–PAGE gel electrophoresis was performed using the Laemmli method. Samples on the gel were transferred to an Immobilon-P polyvinylidene fluoride (PVDF) membrane (Millipore) for ∼2 h using a semidry transfer system. Primary antibody solutions were composed of 1X TBS, 0.1% Tween-20, 5% dry milk (wt/vol), 0.02% NaN3, and antibody. The primary antibodies for tagged protein detection in this study were c-Myc (mouse 9E10, Santa Cruz Biotechnology, used at 1:1000) and HA (mouse anti-HA.11, Covance, used at 1:1000). Cdc28 and Pho85 served as cell-cycle loading controls and were detected with mouse anti-PSTAIR (mouse, Abcam, used at 1:10,000 or 1:16,667). Secondary antibodies were horseradish peroxidase–conjugated (horse anti-mouse immunoglobulin G, Cell Signaling Technology, used at 1:3000). Blots were visualized with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific). Immunoblot quantification and normalization to the loading control were performed in ImageJ.
Protein sample preparation for mass spectrometry
Stable isotope-labeled (SIL) peptides (SpikeTide TQL) were ordered from JPT Peptide Technologies (Berlin, Germany) and were digested with trypsin as previously described (Foster et al., 2014). Three peptides per protein of interest were selected based on available data, preferring 1) peptides utilized in a prior targeted study of yeast TFs (Mirzaei et al., 2013) and 2) peptides with optimal characteristics for targeted proteomics that had been identified in discovery-based proteomics studies (www.peptideatlas.org/; Desiere et al., 2006). In the absence of these data, we selected proteotypic peptides as previously designed and analyzed by Aebersold and coworkers (Picotti et al., 2013).
Yeast protein samples were isolated as described above. TCA pellets were resuspended in 100 µl of 0.25% (wt/vol) acid labile surfactant (ALS-1) in 50 mM ammonium bicarbonate, pH 8 (AmBic), followed by probe sonication for 4 × 3 s and shaking at 50°C for 10 min. After centrifugation at 15,000 ×g for 20 min, protein concentrations of supernatants were measured by Bradford assay. Protein (20 µg/sample) was reduced and denatured by heating at 80°C for 10 min in buffer containing 0.2% ALS-1 and 10 mM DTT. After cooling, samples were alkylated with 25 mM iodoacetamide for 30 min and digested overnight with 1:50 (wt/wt) of sequencing grade modified trypsin (Promega) at 37°C. Following digestion, 2% (vol/vol) acetonitrile and 1% (vol/vol) TFA were added and the digests were incubated at 60°C for 2 h. After centrifugation, SIL peptides (Supplemental Table 1) were added to a final concentration of 5 fmol of each peptide per µg of peptide digests. Finally, peptides were transferred to maximum recovery LC vials (Waters).
Quantitative LC-MS/MS
Targeted quantitation of yeast proteins was performed using parallel reaction monitoring (PRM; Gallien et al., 2012, 2014). Briefly, 1 µg of peptide digests per sample was analyzed using a nanoACQUITY UPLC system (Waters) coupled to a QExactive Plus high-resolution accurate mass tandem mass spectrometer (Thermo) via a nanoelectrospray ionization source. Peptides were trapped on a Symmetry C18 180 µm × 20 mm trapping column (5 μl/min at 99.9/0.1 vol/vol H2O/MeCN) followed by analytical separation using a 1.7-µm ACQUITY HSS T3 C18 75 µm × 250 mm column (Waters) with a 90-min gradient of 5–40% MeCN with 0.1% formic acid at a flow rate of 400 nl/min and column temperature 55°C. Data collection on the QExactive Plus MS was performed in targeted MS/MS mode at resolution 17,500 (m/z 200) with a target AGC value of 5 × 104 ions, an isolation width of 1.0 m/z, and an ion fill time of 240 ms. Targeted MS/MS was triggered by an inclusion list, with 2-min retention time windows for each precursor.
PRM data analysis
PRM data were analyzed using Skyline (MacLean et al., 2010). To generate a spectral library, 50 fmol of neat SpikeTides were analyzed by data-dependent LC-MS/MS using nanoACQUITY and QExactive Plus MS followed by database searching using Mascot v2.5. Raw MS/MS data were imported into Skyline using the following transition setting tabs: filter, y-ions, >precursor m/z to last ion; and Full-scan, targeted acquisition using an Orbitrap analyzer with 17,500 resolution @ 200 m/z. Default peptide settings were used except for the structural modification, carbamidomethyl-Cys, and the isotope modifications, label: 13C615N4-Arg and label: 13C615N2-Lys isotope. Curated Skyline files and raw data were uploaded to the Panorama Targeted Proteomics data repository (panoramaweb.org; Sharma et al., 2014) and can be accessed at https://panoramaweb.org/yeast_cell_cycle_prm.url. Data can also be accessed via ProteomeXchange (PXD010937). Normalized PRM data from this study are available in Supplemental Tables 6 and 7.
TAKT algorithm details
The Kendall tau (KT) distance (Kendall, 1938) may be thought of as endowing a group of rankings of a fixed list of length n with a metric structure: the distance between two rankings is taken to be the number of pairs of ranks that are in a different order in the two rankings. Because there are n(n-1)/2 possible pairs in a list of length n, a normalized KT distance is obtained by simply dividing the number of pairs of ranks that are in a different order in the two rankings by the total possible number of these so-called discordances. The KT metric can extend to a dissimilarity measure between partial rankings—generated by lists with repeated values—by also counting as discordances those instances in which the pair is ambiguously ranked in one ranking but fully ordered in the other ranking (Fagin et al., 2006).
We consider a gene expression profile to be a (partial) ranking of the time points at which measurements were made in a time-course experiment. In other words, we assign to the list of n time points a (partial) ranking according to the measured expression levels, assigning rank 1 to the time at which the minimum expression was attained, and the time of the maximum measurement the rank m≤n (allowing for the possibility of ambiguous rankings due to repeated expression levels). By using the KT distance to compare the (partial) rankings—determined by two different expression profiles, v = (v(t1), v(t2), …, v(tn)) and w = (w(t1), w(t2), …, w(tn))—of the list of times points at which measurements were made, we record the number of pairs of time points {ti, tj} for which there is a discordancy in the rankings of these time points, i.e., the number of instances where v(ti) < v(tj) while w(ti) > w(tj) or v(ti) > v(tj) while w(ti) < w(tj), or those instance where v(ti) = v(tj) while w(ti) ≠ w(tj), or vice versa. In this way, the KT measure of dissimilarity captures disagreement in the “up” and “down”’ patterns of expression between pairs of time points.
The KT metric has been used elsewhere to study the dynamic properties of time-series gene expression data. In particular, the periodicity detection algorithm JTK_CYCLE uses the KT metric to measure the disagreement in the “up” and “down” patterns of expression between a measured gene expression time course and template profiles from known periodic functions (Hughes et al., 2010).
Because the KT metric depends only on the ranking of time points by an expression profile, it is invariant to certain transformations of an expression profile; in other words, the distance between the curves v and w is the same as the distance between v and bw + c, where c is any constant and b is any positive constant. This property is crucial when trying to compare the dynamics between two fundamentally different quantities whose units are not comparable, such as the various protein and transcript abundance measurements reported here. Furthermore, even profiles that are dynamically identical but are shifted in time (i.e., v(t) = w(t + c) for all t, for some c) may be considered dissimilar by the KT metric. For this reason, the TAKT algorithm minimizes the normalized KT dissimilarity measure across shifts of the time points of one curve (through one cell-cycle period) while holding the other curve fixed, and reports both the original dissimilarity—which may be large due only to a temporal delay—and the minimal dissimilarity across time shifts. The time shift that yields the minimal normalized KT measure of dissimilarity is then defined as the estimate of the time delay between mRNA and protein expression.
To determine the significance of the TAKT measure of dissimilarity between a protein abundance profile and its corresponding mRNA profile, 10,000 random mRNA abundance profiles were generated for each mRNA curve by permuting its time points, and the TAKT dissimilarity was computed between each random curve and the fixed protein profile. The fraction of pairs whose dissimilarity is not larger than the dissimilarity between the true profiles is then reported as an empirical p value.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chun-Yi Cho for helpful discussions and manuscript suggestions. This work is supported by Defense Advanced Research Projects Agency (DARPA) Grant D12AP00025.
Abbreviations used:
- APC/C
anaphase-promoting complex/cyclosome
- CDK
cyclin-dependent kinase
- DTT
dithiothreitol
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- MBF
Mbp1/Swi6
- PRM
parallel reaction monitoring
- PVDF
polyvinylidene fluoride
- SBF
Swi4/Swi6
- SCF
Skp1/Cul1/F-box
- SFF
Swi Five Factor
- SIL
stable isotope-labeled
- TAKT
Temporal Alignment Kendall-Tau
- TCA
trichloroacetic acid
- TFs
transcription factors
- YEPD
yeast extract–peptone–dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-04-0255) on September 12, 2018.
REFERENCES
- Amon A, Tyers M, Futcher B, Nasmyth K. (1993). Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell , 993–1007. [DOI] [PubMed] [Google Scholar]
- Ball DA, Adames NR, Reischmann N, Barik D, Franck CT, Tyson JJ, Peccoud J. (2013). Measurement and modeling of transcriptional noise in the cell cycle regulatory network. Cell Cycle , 3203–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball DA, Marchand J, Poulet M, Baumann WT, Chen KC, Tyson JJ, Peccoud J. (2011). Oscillatory dynamics of cell cycle proteins in single yeast cells analyzed by imaging cytometry. PLoS One , e26272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C, Skotheim JM, de Bruin RAM. (2013). Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol , 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. (2010). A global protein kinase and phosphatase interaction network in yeast. Science , 1043–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpy A, Krug K, Graf S, Koch A, Popic S, Hauf S, Macek B. (2014). Absolute proteome and phosphoproteome dynamics during the cell cycle of fission yeast. Mol Cell Proteomics , 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C, Motta F, Kelliher C, Deckard A, Haase SB. (2017a). Reconciling conflicting models for global control of cell-cycle transcription. Cell Cycle , 1965–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CY, Kelliher CM, Haase SB. (2017b). The cell-cycle transcriptional network generates and transmits a pulse of transcription once each cell cycle. BioRxiv, https://doi.org/10.1101/190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong YT, Koh JL, Friesen H, Duffy SK, Cox MJ, Moses A, Moffat J, Boone C, Andrews BJ. (2015). Yeast proteome dynamics from single cell imaging and automated analysis. Cell , 1413–1424. [DOI] [PubMed] [Google Scholar]
- Costenoble R, Picotti P, Reiter L, Stallmach R, Heinemann M, Sauer U, Aebersold R. (2011). Comprehensive quantitative analysis of central carbon and amino-acid metabolism in Saccharomyces cerevisiae under multiple conditions by targeted proteomics. Mol Syst Biol , 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin RAM, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J, 3rd, Russell P, Wittenberg C. (2006). Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell , 483–496. [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M. (2008). Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature , 1251–1254. [DOI] [PubMed] [Google Scholar]
- Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R. (2006). The PeptideAtlas project. Mol Syst Biol (Database issue), D655–D658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Talia S, Wang H, Skotheim JM, Rosebrock AP, Futcher B, Cross FR. (2009). Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol , e1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Spottswood MR. (1991). A new human p34 protein kinase, CDK2, identified by complementation of a cdc28 mutation in Saccharomyces cerevisiae, is a homolog of Xenopus Eg1. EMBO J , 2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott SG, McLaughlin CS. (1978). Rate of macromolecular synthesis through the cell cycle of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA , 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. (2011). Commitment to a cellular transition precedes genome-wide transcriptional change. Mol Cell , 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagin R, Kumar R, Mahdian M, Sivakumar D, Vee E. (2006). Comparing partial rankings. SIAM J Discrete Math , 628–648. [Google Scholar]
- Flory MR, Lee H, Bonneau R, Mallick P, Serikawa K, Morris DR, Aebersold R. (2006). Quantitative proteomic analysis of the budding yeast cell cycle using acid-cleavable isotope-coded affinity tag reagents. Proteomics , 6146–6157. [DOI] [PubMed] [Google Scholar]
- Foster MW, Thompson JW, Ledford JG, Dubois LG, Hollingsworth JW, Francisco D, Tanyaratsrisakul S, Voelker DR, Kraft M, Moseley MA, Foster WM. (2014). Identification and quantitation of coding variants and isoforms of pulmonary surfactant protein A. J Proteome Res , 3722–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. (1999). A sampling of the yeast proteome. Mol Cell Biol , 7357–7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S, Bourmaud A, Kim SY, Domon B. (2014). Technical considerations for large-scale parallel reaction monitoring analysis. J Proteomics , 147–159. [DOI] [PubMed] [Google Scholar]
- Gallien S, Duriez E, Crone C, Kellmann M, Moehring T, Domon B. (2012). Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol Cell Proteomics , 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Wells GP, Smerdon SJ, Sedgwick SG. (2004). Phosphorylation status and cellular localization of Swi6 Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol Cell Biol , 2277–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. (2003). Global analysis of protein expression in yeast. Nature , 737–741. [DOI] [PubMed] [Google Scholar]
- Goranov AI, Cook M, Ricicova M, Ben-Ari G, Gonzalez C, Hansen C, Tyers M, Amon A. (2009). The rate of cell growth is governed by cell cycle stage. Genes Dev , 1408–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GD, Brooks L, Zhang X, Mahoney JM, Martyanov V, Wood TA, Sherlock G, Cheng C, Whitfield ML. (2013). Identification of cell cycle–regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol Biol Cell , 3634–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Bernard A, Orlando DA, Haase SB, Hartemink AJ. (2013). Branching process deconvolution algorithm reveals a detailed cell-cycle transcription program. Proc Natl Acad Sci USA , E968–E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. (1999). Correlation between protein and mRNA abundance in yeast. Mol Cell Biol , 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Reed SI. (1999). Evidence that a free running oscillator drives G1 events in the budding yeast cell cycle. Nature , 394–397. [DOI] [PubMed] [Google Scholar]
- Haase SB, Wittenberg C. (2014). Topology and control of the cell-cycle-regulated transcriptional circuitry. Genetics , 65–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Warren EN, Borchers CH. (2004). Multi-kinase phosphorylation of the APC/C activator Cdh1 revealed by mass spectrometry. Cell Cycle , 1278–1284. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. (1974). Genetic control of the cell division cycle in yeast. Science , 46–51. [DOI] [PubMed] [Google Scholar]
- Hillenbrand P, Maier KC, Cramer P, Gerland U. (2016). Inference of gene regulation functions from dynamic transcriptome data. eLife , e12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak CE, Luscombe NM, Qian J, Bertone P, Piccirrillo S, Gerstein M, Snyder M. (2002). Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev , 3017–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JN, Park I, Ellingson E, Littlepage LE, Pellman D. (2001). Activity of the APC Cdh1 form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. J Cell Biol , 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. (2010). JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms , 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. (2003). Global analysis of protein localization in budding yeast. Nature , 686–691. [DOI] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. (2001). Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol , 4684–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. (2001). Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature , 533–538. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Jensen TS, de Lichtenberg U, Brunak S, Bork P. (2006). Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature , 594–597. [DOI] [PubMed] [Google Scholar]
- Kelliher CM, Leman AR, Sierra CS, Haase SB. (2016). Investigating conservation of the cell-cycle-regulated transcriptional program in the fungal pathogen, Cryptococcus neoformans. PLoS Genet , e1006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M. (1938). A new measure of rank correlation. Biometrika , 81–93. [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. (2014). Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods , 319–324. [DOI] [PubMed] [Google Scholar]
- Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bähler J. (2007). A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell , 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry BD, Mapa CE, Arsenault HE, Poti KE, Benanti JA. (2014). Regulation of a transcription factor network by Cdk1 coordinates late cell cycle gene expression. EMBO J , 1044–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Nurse P. (1987). Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature , 31–35. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. (2002). Transcriptional regulatory networks in Saccharomyces cerevisiae. Science , 799–804. [DOI] [PubMed] [Google Scholar]
- Leman AR, Bristow SL, Haase SB. (2014). Analyzing transcription dynamics during the budding yeast cell cycle. Methods Mol Biol , 295–312. [DOI] [PubMed] [Google Scholar]
- Li JJ, Bickel PJ, Biggin MD. (2014). System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ , e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord PG, Wheals AE. (1981). Variability in individual cell cycles of Saccharomyces cerevisiae. J Cell Sci , 361–376. [DOI] [PubMed] [Google Scholar]
- Ly T, Ahmad Y, Shlien A, Soroka D, Mills A, Emanuele MJ, Stratton MR, Lamond AI. (2014). A proteomic chronology of gene expression through the cell cycle in human myeloid leukemia cells. eLife , e01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. (2010). Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics , 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bähler J. (2012). Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell , 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoff KA, Guo X, Deckard A, Kelliher CM, Leman AR, Francey LJ, Hogenesch JB, Haase SB, Harer JL. (2016). The local edge machine: inference of dynamic models of gene regulation. Genome Biol , 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JAH. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J , 546–566. [DOI] [PubMed] [Google Scholar]
- Mirzaei H, Knijnenburg TA, Kim B, Robinson M, Picotti P, Carter GW, Li S, Dilworth DJ, Eng JK, Aitchison JD, et al. (2013). Systematic measurement of transcription factor-DNA interactions by targeted mass spectrometry identifies candidate gene regulatory proteins. Proc Natl Acad Sci USA , 3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J, Nomoto S, Yasuda H, Reed SI, Matsumoto K. (1991). Cloning of a human cDNA encoding a CDC2-related kinase by complementation of a budding yeast cdc28 mutation. Proc Natl Acad Sci USA , 9006–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Iversen ES, Hartemink AJ, Haase SB. (2007). A probabilistic model for cell cycle distributions in synchrony experiments. Cell Cycle , 478–488. [DOI] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Wang JY, Socolar JES, Iversen ES, Hartemink AJ, Haase SB. (2008). Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature , 944–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, O’Connell JD, Gaun A, Gygi SP. (2015). Proteome-wide quantitative multiplexed profiling of protein expression: carbon-source dependency in Saccharomyces cerevisiae. Mol Biol Cell , 4063–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. (2009). Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell , 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Clément-Ziza M, Lam H, Campbell DS, Schmidt A, Deutsch EW, Röst H, Sun Z, Rinner O, Reiter L, et al. (2013). A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature , 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T, Miles S, GuhaThakurta D, Jemiolo D, Breeden LL. (2002). Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev , 3034–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T, Wu W, Miles S, Noble WS, Breeden LL. (2006). The forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev , 2266–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahal R, Amon A. (2008). Mitotic CDKs control the metaphase-anaphase transition and trigger spindle elongation. Genes Dev , 1534–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds D, Shi BJ, McLean C, Katsis F, Kemp B, Dalton S. (2003). Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires C1b kinase activity: a mechanism for CLB cluster gene activation. Genes Dev , 1789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. (2000). Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol , 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustici G, Mata J, Kivinen K, Lió P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bähler J. (2004). Periodic gene expression program of the fission yeast cell cycle. Nat Genet , 809–817. [DOI] [PubMed] [Google Scholar]
- Serikawa KA, Xu XL, MacKay VL, Law GL, Zong Q, Zhao LP, Bumgarner R, Morris DR. (2003). The transcriptome and its translation during recovery from cell cycle arrest in Saccharomyces cerevisiae. Mol Cell Proteomics , 191–204. [DOI] [PubMed] [Google Scholar]
- Sevim V, Gong X, Socolar JES. (2010). Reliability of transcriptional cycles and the yeast cell-cycle oscillator. PLoS Comput Biol , e1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Eckels J, Taylor GK, Shulman NJ, Stergachis AB, Joyner SA, Yan P, Whiteaker JR, Halusa GN, Schilling B, et al. (2014). Panorama: a targeted proteomics knowledge base. J Proteome Res , 4205–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons Kovacs LA, Mayhew MB, Orlando DA, Jin Y, Li Q, Huang C, Reed SI, Mukherjee S, Haase SB. (2012). Cyclin-dependent kinases are regulators and effectors of oscillations driven by a transcription factor network. Mol Cell , 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, Young RA. (2001). Serial regulation of transcriptional regulators in the yeast cell cycle. Cell , 697–708. [DOI] [PubMed] [Google Scholar]
- Soste M, Hrabakova R, Wanka S, Melnik A, Boersema P, Maiolica A, Wernas T, Tognetti M, von Mering C, Picotti P. (2014). A sentinel protein assay for simultaneously quantifying cellular processes. Nat Methods , 1045–1048. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. (1998). Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell , 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villén J. (2013). Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature , 859–864. [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet , 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Koller A, Oshiro G, Ulaszek RR, Plouffe D, Deciu C, Winzeler E, Yates JR., 3rd (2003). Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc Natl Acad Sci USA , 3107–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong FM, Lim HH, Wang Y, Surana U. (2001). Early expressed Clb proteins allow accumulation of mitotic cyclin by inactivating proteolytic machinery during S phase. Mol Cell Biol , 5071–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yofe I, Weill U, Meurer M, Chuartzman S, Zalckvar E, Goldman O, Ben-Dor S, Schütze C, Wiedemann N, Knop M, et al. (2016). One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat Methods , 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Srividhya J, De Luca T, Lee JH, Pomerening JR. (2014). Uncovering the role of APC-Cdh1 in generating the dynamics of S-phase onset. Mol Biol Cell , 441–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. (1998). Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science , 1721–1724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.