Abstract
Centrosomes are the major microtubule-nucleating and microtubule-organizing centers of cells and play crucial roles in microtubule anchoring, organelle positioning, and ciliogenesis. At the centrosome core lies a tightly associated or “engaged” mother–daughter centriole pair. During mitotic exit, removal of centrosomal proteins pericentrin and Cep215 promotes “disengagement” by the dissolution of intercentriolar linkers, ensuring a single centriole duplication event per cell cycle. Herein, we explore a new mechanism involving vesicular trafficking for the removal of centrosomal Cep215. Using small interfering RNA and CRISPR/Cas9 gene-edited cells, we show that the endocytic protein EHD1 regulates Cep215 transport from centrosomes to the spindle midbody, thus facilitating disengagement and duplication. We demonstrate that EHD1 and Cep215 interact and show that Cep215 displays increased localization to vesicles containing EHD1 during mitosis. Moreover, Cep215-containing vesicles are positive for internalized transferrin, demonstrating their endocytic origin. Thus, we describe a novel relationship between endocytic trafficking and the centrosome cycle, whereby vesicles of endocytic origin are used to remove key regulatory proteins from centrosomes to control centriole duplication.
INTRODUCTION
The centrosome serves as the major microtubule-organizing center of a cell, giving rise to a variety of protein machines that include mitotic spindles and cilia (Conduit et al., 2015). Centrosome duplication is tightly coupled to the cell cycle and normally occurs only once each cycle. Typically, cells in G1 phase contain a single centrosome. During S phase, centrosomes duplicate, with mother and daughter centrioles spawning an orthogonally positioned procentriole. As newly formed daughter centrioles elongate during G2 phase, the protein linker connecting the two centrosomes is broken, triggering centrosome separation (Bahe et al., 2005). Centrosomes then recruit additional pericentriolar material (PCM) in a process called “maturation,” which allows them to facilitate spindle assembly. During mitotic exit, each daughter cell receives a tightly associated mother–daughter centriole pair that disconnects in a process termed “disengagement” (Tsou and Stearns, 2006a; Mardin and Schiebel, 2012). Thus, the centrosome cycle ensures that mitotic cells contain two centrosomes to guide assembly of a bipolar spindle and enhance the accuracy of chromosome segregation.
To prevent rampant centrosome amplification, the centrosome cycle contains roadblocks that restrict duplication and that are normally removed at distinct steps during the cell cycle. For example, after assembly of a new daughter centriole, pericentrin (PCNT) maintains the orthogonal “engaged” configuration of the mother–daughter pair. Importantly, engagement itself functions as a block to centriole reduplication (Tsou and Stearns, 2006b). During mitotic exit, PCNT is cleaved to promote disengagement and “license” centrioles for another round of duplication (Lee and Rhee, 2012). Equally crucial is the removal of the PCM protein Cep215/Cdkrap2 from centrosomes. Notably, PCNT interacts with Cep215 and is required for its centrosomal localization (Pagan et al., 2015). Although expression of a noncleavable PCNT mutant blocks centriole disengagement (and thus duplication), the depletion of Cep215 rescues centriole duplication in these cells. Therefore, in addition to PCNT cleavage, Cep215 is normally removed from centrosomes for centriole disengagement and duplication to proceed in a timely manner.
At present, little is known of how centrosomal proteins are recruited and removed from centrosomes. Although proteasome-mediated degradation is a mechanism for eliminating centrosomal proteins (Chen et al., 2002; Pagan et al., 2015), it is assumed that diffusion accounts for the exchange of most centrosomal proteins. However, an unexplored possibility is that membrane trafficking assists protein removal. Indeed, several proteins, including ninein, undergo relocation from centrosomes to the spindle midbody to promote centrosome duplication (Bouckson-Castaing et al., 1996). Moreover, ninein moves from centrosomes along microtubules, suggesting it may be redistributed via a transport mechanism (Moss et al., 2007).
Here, we explore vesicular trafficking as a mechanism to attenuate the levels of centrosomal proteins and their redistribution to the spindle midbody. Our findings suggest a role for the endocytic ATPase protein EHD1, its interaction partner MICAL-L1, and the retromer complex in the removal and transport of Cep215 from the centrosome to the spindle midbody, thus enabling centriole disengagement and duplication. Our data implicate a previously unknown vesicular trafficking pathway in the regulation of the centrosome cycle.
RESULTS AND DISCUSSION
Depletion of EHD1 and select endocytic regulatory proteins impairs centriole disengagement and duplication
Several studies have linked endocytic trafficking regulators to various aspects of centrosome biology. For example, CP110 is removed from mother centrioles to promote ciliogenesis, and this requires both ciliary vesicles and the endocytic regulatory protein EHD1 (Lu et al., 2015). Furthermore, Rab11-associated recycling endosomes interact with mother centriole appendages and contribute to mitotic spindle assembly (Hehnly and Doxsey, 2014). In spite of these discoveries, a potential role for endocytic transport in regulation of the centrosome cycle has not been explored.
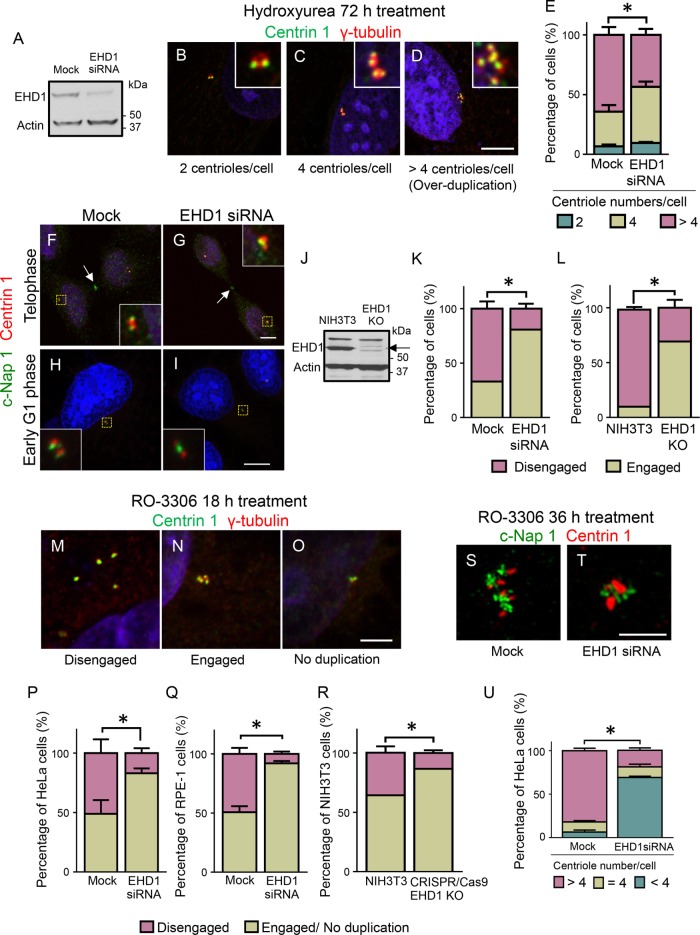
We first examined whether EHD1 might participate in centriole duplication using an established centriole reduplication assay in U2OS cells. Specifically, hydroxyurea (HU) was added to cells to induce a prolonged S-phase arrest, which causes repeated cycles of mother–daughter centriole disengagement and duplication (Kuriyama et al., 2007). Centriole numbers were measured in mock-treated and EHD1-depleted cells by immunostaining for γ-tubulin and Centrin 1. EHD1 protein levels were depleted by 75–85% as determined by anti-EHD immunoblotting (Figure 1A). Whereas Centrin 1 labels all centriole distal tips, γ-tubulin appears as a single spot associated with either unduplicated (Figure 1B) or duplicated centriole pairs (Figure 1C) and individual centriole singlets (Figure 1D). Although most control cells contained >4 centrioles, the percentage of cells with >4 centrioles was significantly reduced in EHD1-depleted cells (Figure 1E). These findings suggest that EHD1, an endocytic membrane-remodeling protein, plays an unexpected role in centriole duplication.
FIGURE 1:
Centrosome duplication and centriole disengagement are impaired upon EHD1 depletion. (A) Immunoblotting demonstrates the efficiency (75–85%) of siRNA-mediated EHD1 depletion from U2OS cells. Equal total protein amounts were loaded. Actin, loading control. (B–D) U2OS cells were HU treated for 72 h, immunostained for Centrin 1 (green) and γ-tubulin (red), and imaged by confocal microscopy. Representative examples of cells containing 2 centrioles (B), 4 centrioles (C), and > 4 centrioles (D). DNA, blue. Insets show centrioles at higher magnification. Scale bar: 5 μm (in all images). (E) Graph shows average percentages of mock-treated and EHD1-depleted cells containing the indicated number of centrioles. For all graphs, n = 3 experiments (>100 cells quantified per each experiment). Error bars indicate SD. *, p < 0.0001. (F–I) Centriole disengagement is impaired upon EHD1 depletion. HeLa cells were mock treated (F, H) or EHD1 depleted (G, I), synchronized to telophase (F, G) or early G1 phase (H, I), and immunostained for C-Nap 1 (green) and Centrin 1 (red). Cells with two c-Nap1 foci contain disengaged centrioles (F, H), while cells with one c-Nap1 spot contain an engaged centriole pair (G, I). DNA, blue. Insets show centrioles at higher magnification. (J) The level of EHD1 in wild-type or CRISPR/Cas9 EHD1 knockout NIH 3T3 cells was determined by anti-EHD1 immunoblot (arrow denotes EHD1). Actin, loading control. (K, L) Graph shows the percentage of cells with disengaged/engaged centrioles in late cytokinesis siRNA-treated U2OS (K) and CRISPR/Cas9 gene-edited EHD1-knockout NIH 3T3 cells (L). (M–R) Premature centriole disengagement induced by the Cdk1 inhibitor RO-3306 is impaired in EHD1-depleted cells. HeLa and RPE-1 cells were treated with RO-3306 for 18 h and immunostained for γ-tubulin (red) and Centrin 1 (green). Micrographs show representative HeLa cells with disengaged centrioles (M), engaged centrioles (N), or centrioles that fail to duplicate (O). DNA, blue. (P–R) Graphs show the percentage of cells containing either disengaged centrioles or an engaged/no duplication phenotype in RO-3306 and siRNA-treated HeLa cells (P), RPE-1 cells (Q), and wild-type NIH 3T3 and CRISPR/Cas9 EHD1-knockout cells (R). (S–U) Reduplication of disengaged centrioles is impaired in EHD1-depleted cells. Mock-treated or EHD1-depleted HeLa cells were incubated with RO-3306 for 36 h and immunostained for c-Nap1 (green) and Centrin 1 (red). The percentage of cells with >4 centrioles (S) or 4 centrioles or <4 centrioles (T) is shown in graph (U). Asterisks denote statistical significance between mock-treated and EHD1-depleted cells with >4 centrioles.
We next examined whether this centriole reduplication defect could be due to an inability of mother–daughter centriole pairs to disengage, because failure to disengage prevents centriole duplication (Tsou and Stearns, 2006a). We synchronized control or EHD1-depleted cells and examined their centriole engagement status during telophase and early G1 phase by immunostaining for c-Nap1 and Centrin 1. After disengagement, c-Nap1 binds the proximal ends of both mother and daughter centrioles, appearing as two distinct foci (Fry et al., 1998). However, within an engaged pair, c-Nap1 localizes as a single spot to the proximal end of the mother centriole (Tsou and Stearns, 2006b). Thus, disengaged centrioles contain a single spot of c-Nap1 and Centrin 1 (1:1 ratio), whereas an engaged pair has one focus of c-Nap1 and two Centrin 1 spots (1:2 ratio) (Tsou and Stearns, 2006b). As expected, the majority of centrioles (65%) were disengaged in control (Mock) cells (Figure 1, F, H, and K). However, upon EHD1 depletion, less than 20% of cells contained disengaged centrioles (Figure 1, G, I, and K), suggesting that EHD1 is required for efficient centriole disengagement.
As an alternative approach to small interfering RNA (siRNA), we used CRISPR/Cas9 gene editing to knockout EHD1 (EHD1 KO) in NIH 3T3 cells (Yeow et al., 2017) (Figure 1J) and measured the percentage of cells with disengaged centrioles compared with the wild-type NIH 3T3 line. Strikingly, we found a decrease in the percentage of EHD1 knockout cells with disengaged centrioles, indicating that the effect on centrioles is not limited to acute EHD1 depletion (Figure 1L). These data indicate that EHD1 plays a significant role in promoting centriole disengagement.
We tested this idea further using an alternative centriole disengagement assay. Cells were treated with the Cdk1 inhibitor, RO-3306, which causes G2-phase arrest and promotes the premature disengagement of duplicated centriole pairs (Vassilev et al., 2006; Prosser et al., 2012). Centriole disengagement was visualized by immunostaining for γ-tubulin and Centrin 1. Accordingly, γ-tubulin:Centrin 1 foci in a 4:4 ratio indicates premature disengagement (Figure 1M), a 4:2 ratio represents two pairs of engaged centrioles (Figure 1N), and a 2:1 ratio signifies an unduplicated centriole pair (Figure 1O). Whereas most control cells (52%) displayed premature centriole disengagement (4:4 ratio), this was reduced (22%) in EHD1-depleted HeLa cells (Figure 1P). Premature centriole disengagement was also decreased in EHD1-depleted immortalized RPE-1 cells (Figure 1Q) and NIH 3T3 CRISPR/Cas9 EHD1 knockout cells (Figure 1R). In addition, we transiently transfected green fluorescent protein (GFP)-EHD1 into either the wild-type NIH 3T3 cells (Supplemental Figure 1, A–C) or the EHD1 knockout cells (Supplemental Figure 1, D–F) and calculated the percentages of cells with engaged or disengaged centrioles (Supplemental Figure 1G). Whereas the EHD1 knockout cells had a higher percentage of engaged centrioles than wild-type NIH 3T3 or GFP-EHD1–transfected NIH 3T3 cells, transfection of the knockout cells restored the level of cells with engaged centrioles to those of the wild-type NIH 3T3 cells (Supplemental Figure 1G).
We also found that prolonged treatment with RO-3306, which normally promotes reduplication of disengaged centrioles in HeLa cells (Loncarek et al., 2010; Steere et al., 2011) (Figure 1, S and U), does not have this effect in EHD1-depleted cells, the majority of which contained fewer than 4 centrioles (Figure 1, T and U). Collectively, the participation of EHD1 in centriole disengagement implicates vesicular transport as a novel component of the centrosome cycle.
Cep215 and PCNT are trafficked from centrosomes to the midbody in an EHD1-dependent manner
How could vesicular transport impact centriole duplication? One possibility is that vesicular transport is used as a mechanism to remove centrosomal proteins that normally restrict progression through the centrosome cycle. Cep215 is an excellent candidate for such a mechanism, as its presence on centrosomes is a barrier to centriole duplication (Graser et al., 2007; Barrera et al., 2010; Pagan et al., 2015). Cep215 also associates with microtubules and, in addition to its centrosomal localization, appears in a punctate cytoplasmic pattern (Fong et al., 2008, 2009). However, little is known about the dynamics of these punctate structures and their relationship to the Cep215 centrosome-bound pool. We therefore hypothesized that EHD1 depletion may cause abnormal centrosomal retention of Cep215. In mock-treated cells, Cep215 fluorescence intensity on centrosomes decreased during mitotic exit, as previously reported (Pagan et al., 2015), with Cep215 partially relocalized to the spindle midbody during cytokinesis (Figure 2, A and C; yellow arrows in C mark Cep215 on the centrosome and rectangular ROI denote Cep215 at the spindle midbody). (Note that cells in Figure 2, C and D, were imaged at a lower intensity compared with cells in Figure 2, A and B, so that differences in Cep215 centrosome localization can be detected, whereas cells in Figure 2, A and B, were imaged at a higher intensity to better quantify Cep215 at the spindle midbody.) However, in EHD1-depleted cells, Cep215/γ-tubulin intensity on centrosomes was significantly higher during cytokinesis and nearly absent on the midbody (Figure 2, B and D–F; yellow arrows in D and rectangular region of interest). We also observed a similar increase in the centrosomal Cep215/γ-tubulin ratio in late mitotic EHD1 knockout cells as compared with control cells (Figure 2G). Strikingly, Cep215 localization to centrosomes was similarly regulated by other endocytic regulatory proteins, including the EHD1 interaction partner MICAL-L1, as well as components of the retromer complex (Vps26 and Vps35), but not Rab11 (Supplemental Figure 2).
FIGURE 2:
EHD1 regulates Cep215 and PCNT levels on centrosomes and mediates Cep215 recruitment to the midbody during cytokinesis. (A–G) Cep215 accumulates on centrosomes upon EHD1 depletion. HeLa cells were mock treated or EHD1 depleted, synchronized to cytokinesis, and immunostained for Cep215. Ratiometric immunofluorescence intensity of Cep215 (compared with γ-tubulin) on the centrosome (yellow arrows) is significantly higher in EHD1-depleted cells (quantified in E). Cep215 is localized to the midbody in mock-treated cells (A, C, dashed rectangles), but fails to target the midbody in the absence of EHD1 (B, D, dashed rectangles, quantified in F). The ratiometric level of Cep215/γ-tubulin on centrosomes of wild-type NIH 3T3 and CRISPR/Cas9-edited EHD1 knockout cells during cytokinesis is quantified in G. (H–L) PCNT accumulates on centrosomes upon EHD1 depletion. HeLa cells were mock and EHD1-siRNA treated, synchronized to cytokinesis, and immunostained for α-tubulin (H, I) or PCNT (J, K). Immunofluorescence intensity of PCNT on centrosomes (yellow boxes, J, K, and shown at higher magnification in insets) was quantified (L). n = 3 independent experiments, ≥30 cells analyzed per experiment. Error bars are SD. *, p < 0.001. Scale bar: 5 μm.
As with Cep215, PCNT levels also decrease on centrosomes during mitotic exit, an event that coincides with centriole disengagement (Matsuo et al., 2012). Similarly, we found that PCNT accumulates on centrosomes in EHD1-depleted cells (Figure 2, H–L). However, EHD1 depletion does not induce a general redistribution of centrosomal proteins, as both Sas6 (Habedanck et al., 2005) and STIL (Kumar et al., 2009; Stevens et al., 2010) recruitment to centrosomes are unaffected in the absence of EHD1 (Supplemental Figure 3). Overall, these findings support the possibility that EHD1, and several of its endosomal sorting partners, mediates the removal of Cep215 and PCNT from centrosomes.
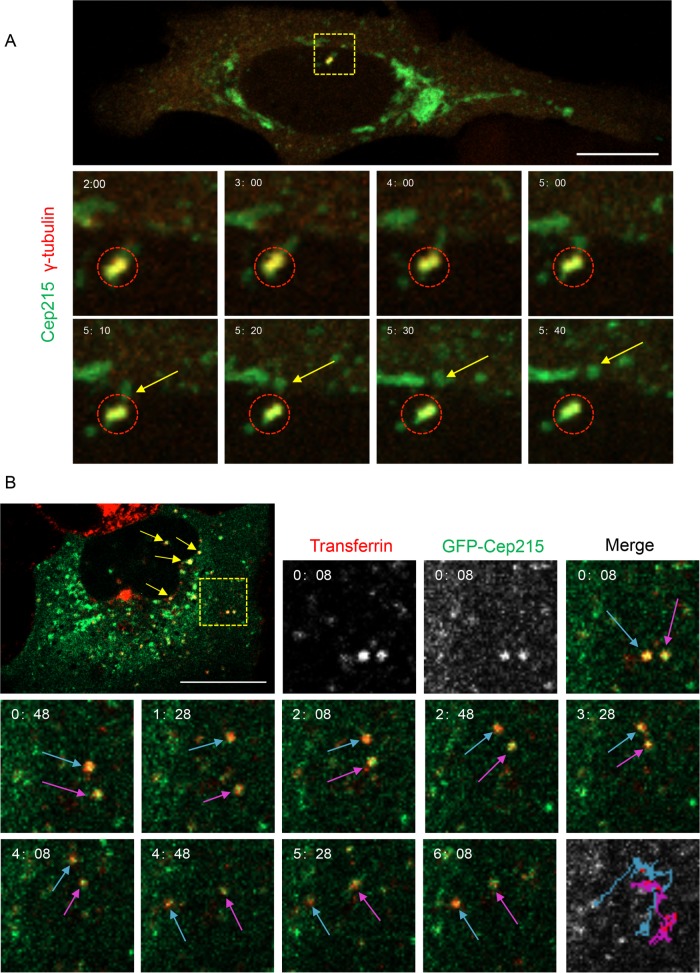
Because global Cep215 protein levels remain constant throughout mitosis (Pagan et al., 2015), the removal of Cep215 from centrosomes is likely due to either diffusion or directed vesicular transport, rather than degradation. Given the role of EHD1 in vesicular transport (Caplan et al., 2002; Gokool et al., 2007; Jovic et al., 2007; Cai et al., 2011), we hypothesized that Cep215’s dynamic localization pattern might occur via EHD-mediated vesicular transport rather than a diffusion-based mechanism. To test this, we used live imaging to investigate whether Cep215 trafficks on vesicles. Cells contained an easily discernible, centrosome-bound population of GFP-Cep215 (Figure 3A, 2 min). Strikingly, Cep215 punctate structures were clearly seen detaching from the centrosome region and moving distally (Figure 3A, yellow arrows, and Supplemental Video 1). Because EHD1 regulates trafficking of a subset of recycling endosomes (Caplan et al., 2002), we next asked whether Cep215 associates with recycling endosomes or markers of the endocytic pathway. To address this question, we imaged GFP-Cep215–expressing live cells pulsed with transferrin, an endocytic cargo whose recycling is governed, in part, by EHD1 (Naslavsky et al., 2004). We noticed multiple events of colocalization between GFP-Cep215 and Alexa Fluor 568–transferrin (Figure 3B, yellow arrows and yellow dashed square; see Supplemental Figure 4 for individual channels at all time points), indicating that a population of Cep215 associates with transferrin-containing recycling endosomes. Importantly, colocalization was not transient, but rather was maintained throughout the experiment (Figure 3B, blue and magenta arrows, and Supplemental Video 2). Moreover, vesicles containing both transferrin and Cep215 were highly mobile (Figure 3B, bottom right panel, blue and magenta tracks depicted over time). These data suggest that a population of Cep215 is trafficked from centrosomes via vesicular transport, possibly through recycling endosomes.
FIGURE 3:
Cep215 is transported away from centrosomes on transferrin-containing vesicles. (A) Cep215 displays directional movement away from the centrosome. U2OS cells transfected with pEGFP-Cep215 and dTomato-γ-tubulin for 16 h were imaged live every 2 s for 10 min. The dashed box indicates the γ-tubulin-labeled centrosome and is shown at higher magnification in the time-stamped insets. Circles mark the centrosome where Cep215 is localized. Arrows point to Cep215 moving away from the centrosome. Scale bar: 10 μm. (B) Cep215 localizes to mobile vesicles containing transferrin. U2OS cells transfected with pEGFP-Cep215 were pulsed for 15 min with Alexa Fluor 568–conjugated transferrin and imaged every 2 s for 10 min. Yellow arrows indicate Cep215 on transferrin-containing vesicles. Insets showcase two vesicles (dashed yellow box) at higher magnification. Blue and magenta arrows mark the movement of these vesicles, and their entire path is illustrated over time (bottom right panel). Videos compiled are representative of 18 (A) or 7 (B) time series obtained. Scale bar: 10 μm.
Video S1.
U2OS cells were transfected with GFP-Cep215 and dTomato-γ-tubulin for 18 h. On the day of imaging, cells were incubated in fresh phenol-red free DMEM and immediately transferred to a heated chamber with 5% CO2 for live cell imaging. Cells were imaged in dual channels every 5 s for 10 min. For each image, 5 z-slices were obtained, each with a 1.5 μm interval. This video demonstrates the z-projection of images at 15 frames per second.
Video S2.
U2OS cells were transfected with GFP-Cep215 for 18 h. On the day of imaging, the cells were incubated in freshly prepared phenol red-free DMEM along with Alexa Fluor568-conjugated transferrin for 15 min. The medium was then replaced with fresh phenol red-free DMEM and cells were transferred to a heated chamber with 5% CO2 for live cell imaging. Images were taken every 2 s for 10 min. The processed video shows images at 15 frames per second.
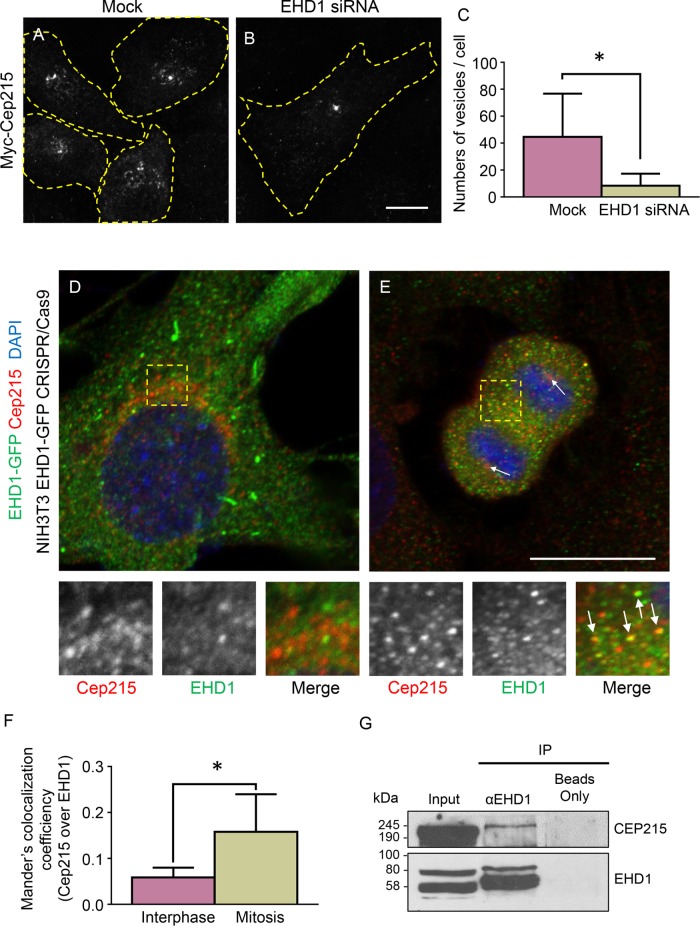
We next examined whether trafficking of Cep215-associated vesicles is mediated by EHD1. Control and EHD-depleted cells were transfected with Myc-Cep215 and immunostained for Myc to identify Cep215-associated vesicular structures (Figure 4, A and B, cell boundaries are marked by dashed yellow regions of interest). Using automated particle detection, we measured the number of Myc-positive vesicles. We found that a significantly smaller number of vesicles were observed in the absence of EHD1 (Figure 4C), possibly due to fewer Cep215-containing vesicles being released into the cytoplasm. Intriguingly, the Cep215 distribution pattern in EHD1-depleted cells was often reminiscent of transferrin-containing endosomes that coalesce at the pericentriolar region (Lin et al., 2001; Giridharan et al., 2013), indicating that EHD1 may regulate the transport of transferrin- and Cep215-associated vesicles in a similar manner.
FIGURE 4:
Cep215 localizes to EHD1-regulated vesicles and forms a complex with EHD1. (A–C) EHD1 regulates the number of Cep215-containing vesicles. Mock-treated and EHD1-depleted U2OS cells were transfected with Myc-Cep215 and immunostained for Myc, and the number of vesicles per cell was calculated (C). Yellow dashed lines mark cell borders. n = 3 independent experiments, 45 cells per experiment quantified. *, p < 0.001. Scale bar: 5 μm. (D–F) Partial colocalization between EHD1 and Cep215 is increased during mitosis. CRISPR/Cas9 gene-edited NIH 3T3 cells expressing EHD1-GFP were fixed by paraformaldehyde and stained with anti-GFP (green) and anti-Cep215 (red). Interphase (D) or anaphase (E) cells were imaged, and the degree of Cep215 colocalization to EHD1-containing structures was analyzed with Mander’s coefficient (F). *, p < 0.001. (G) Cep215 coimmunoprecipitates with EHD1. HeLa cells were lysed and subjected to immunoprecipitations with anti-EHD1 or beads only and then subjected to immunoblotting with anti-Cep215 and anti-EHD1. Five percent of the lysate was included in the inputs. Scale bar: 10 μm.
Cep215 resides in a vesicular complex with EHD1 that is trafficked away from the centrosome
Because EHD1 mediates transport of Cep215-associated vesicles, we examined whether Cep215 could be visualized in vesicles containing EHD1. Using CRISPR/Cas9 gene-edited NIH 3T3 cells containing endogenous levels of EHD1 expressed as an EHD1-GFP fusion protein, we costained interphase and late mitotic cells for Cep215 and 4′,6-diamidino-2-phenylindole (DAPI; Figure 4, D and E). Although some overlap between EHD1- and Cep215-containing structures was detected in interphase cells (Figure 4D, see insets), greater overlap was observed between the two proteins in late mitotic cells (Figure 4E, see insets and arrows). Indeed, a greater than twofold increase in Cep215 that localized to EHD1-containing structures was observed in mitotic cells (Figure 4F). Moreover, in mitotic cells, we visualized endocytic vesicles containing internalized transferrin and both Cep215 and Vps35 in the proximity of the centrosome (Supplemental Figure 5). We therefore rationalized that if EHD1 and Cep215 are transported on common vesicles, then the two proteins may interact with one another. To test this, we performed coimmunoprecipitation experiments. We found that endogenous Cep215 coimmunoprecipitated with endogenous EHD1, whereas Cep215 did not precipitate in the beads-only control (Figure 4G).
Our study shows that EHD1, a regulator of recycling tubule fission, affects centrosome duplication by promoting centriole disengagement. Our findings support a mechanism for centriole disengagement involving the removal of Cep215 from centrosomes using vesicles containing endocytic regulatory proteins such as EHD1, MICAL-L1, and the retromer complex. Moreover, Cep215 trafficks from centrosomes to the spindle midbody during mitotic exit, a time when centriole disengagement occurs. Although separase-mediated PCNT cleavage is a prerequisite for centriole disengagement, a reduction in Cep215 levels on centrosomes is also critical (Pagan et al., 2015). Our results build upon this proteolytic-based pathway for the liberation of a Cep215 pool that resides at the centrosome core. We propose that release of Cep215 from cleaved PCNT is, by itself, not sufficient to promote centriole disengagement but is followed by a vesicular transport step that traffics Cep215 away from centrosomes. In sum, these findings support an unexpected role for vesicular trafficking in the regulation of the centrosome cycle and provide a new mechanism for attenuating the levels of proteins on the organelle.
MATERIALS AND METHODS
Antibodies and reagents
The following antibodies were used: anti-Centrin 1 (Protein Tech), γ-tubulin (Sigma), c-Nap 1 and Sas6 (Santa Cruz), Cep215 (Bethyl & Atlas), PCNT (Abcam), GFP (Roche), EHD1 (Abcam), MICAL-L1 (Novus), Vps26 (Abcam), Vps35 (Abcam), Rab11 (Transduction Laboratories), α-tubulin (Sigma), and actin (Novus). Antibody against STIL was generated in the following manner: Escherichia coli–expressed glutathione-S-transferase (GST)- or maltose-binding protein (MBP)-N-terminal STIL (aa 1148–1287) proteins were purified on either glutathione-Sepharose or amylose resin. Guinea pig polyclonal antisera were raised against purified GST-STIL, and the corresponding MBP fusion was used for antibody affinity purification by precoupling it to Affigel 10/15 resin (Bio-Rad). RO-3306, thymidine, and HU were purchased from Sigma. Transferrin conjugated to Alexa Fluor 568, DAPI, and all secondary antibodies used for immunofluorescence were purchased from Molecular Probes.
Cell culture and treatments
The HeLa cervical cancer cell line (ATCC-CCL2) and the human osteosarcoma cell line U2OS (ATCC-HTB96) were grown at 37°C in 5% CO2 in DMEM (high glucose) (Invitrogen) containing 10% fetal bovine serum (FBS; Sigma), with 2 mM l-glutamine and 100 U/ml penicillin/streptomycin (ThermoFisher). The human epithelial cell line hTERT RPE-1 (ATCC-CRL4000) was cultured at 37°C in 5% CO2 in DMEM/F12 (ThermoFisher) containing 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin/streptomycin, and 1X nonessential amino acids (ThermoFisher). CRISPR/Cas9 was applied to generate the NIH 3T3 cell line expressing endogenous levels of EHD1 with GFP attached to its C-terminus, as well as the EHD1 knockout cells described (Yeow et al., 2017). Wild-type and gene-edited NIH 3T3 cells were cultured at 37°C in 5% CO2 in DMEM containing 10% FBS, with 2 mM l-glutamine and 100 U/ml penicillin/streptomycin. For treatments, cells were plated on collagen (Corning)-coated coverslips.
HeLa and NIH 3T3 cells were synchronized using the double-thymidine block method as previously described with modifications (Ma and Poon, 2017). Cells were treated with 2.5 mM thymidine for 18 h and released into regular media for 9 h, and a second block was performed with 2.5 mM thymidine for 16 h. For collection of cells in S phase, cells were fixed immediately following thymidine treatment; for harvesting of cells in the late cytokinesis stage, thymidine-blocked cells were released into 9 μM RO-3306 for 4 h, washed, and incubated in fresh DMEM for 2 h. For RPE-1 cell synchronization, cells were incubated in DMEM/F12 containing 0.2% FBS for 48 h and then replenished with media containing 10% FBS to trigger their reentry into the cell cycle. Centriole overduplication was induced by 1 mM HU (Calbiochem) treatment on U2OS cells for 72 h. Premature centriole disengagement was triggered by prolonged G2 arrest (Prosser et al., 2012) with the application of 9 μM of the Cdk1 inhibitor RO-3306 for 18 h. RO-3306 treatment was performed for an extended period (36 h) to allow the reduplication of the disengaged centrioles.
Coimmunoprecipitation
HeLa cells growing on 100-mm dishes were lysed in lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 0.5% Triton X-100, iodoacetamide, and freshly added protease inhibitor cocktail. Lysates were cleared by centrifugation at 1889 × g at 4°C for 10 min. The cleared lysate was collected and incubated with anti-EHD1 antibody at 4°C overnight. Protein G agarose beads (GE) were added to the lysate-antibody mix at 4°C for 4 h. Samples were then washed three times with washing buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, iodoacetamide, and 0.1% Triton X-100. Protein complexes were eluted by boiling the sample in the presence of SDS-containing loading buffer and detected by immunoblotting.
DNA constructs, transfection, and siRNA treatment
tdTomato-Gamma-Tubulin-17 was obtained from Addgene (plasmid #58099). Cloning of GFP-Myc-EHD1 has been described (Caplan et al., 2002; Simone et al., 2013). HeLa cells were transfected with FuGENE 6 (Roche Diagnostics) for 18 h at 37°C for live imaging. The GFP-Cep215 construct for live-cell imaging was a generous gift from Robert Z. Qi (Hong Kong University of Science and Technology; Fong et al., 2009).
siRNA oligonucleotides targeting human EHD1 were custom-made by Dharmacon: sense (5′-GAAAGAGAUGCCCAAUGUC(dT)(dT)-3′). siRNA oligonucleotides targeting human Rab11a were purchased from Dharmacon: sense (5′-GAGUAAUCUCCUGUCUCGA(dT)(dT)-3′). Four specific oligonucleotides (On-Target SMART pool, Dharmacon) were directed at human MICAL-L1 (5′-CCGGGUUCCUGGCAAACUA-3′, 5′-GCUAGGAAACAAACGUGAU-3′, 5′-GGUUCAAGCUCAUCCACGA-3′, 5′-GAAUGGGCCUGACGGGCAA-3′), human Vps26a (5′-GCUAGAACACCAAGGAAUU-3′, 5′-UAAAGUGACAAUAGUGAGA-3′, 5′-UGAGAUCGAUAUUGUUCUU-3′, 5′-CCACCUAUCCUGAUGUUAA-3′), or human Vps35 (5′-GAACAUAUUGCUACCAGUA-3′, 5′-GAAAGAGCAUGAGUUGUUA-3′, 5′-GUUGUAAACUGUAGGGAUG-3′, 5′-GAACAAAUUUGGUGCGCCU-3′). HeLa cells were transfected with 100 nM custom-made oligonucleotides targeting EHD1 or Rab11a, or 50 nM On-Target plus SMARTpool oligonucleotides targeting Vps26a, Vps35, or MICAL-L1 using Lipofectamine RNAiMAX (Invitrogen) for 72 h in the absence of antibiotics, as per the manufacturer’s protocol. The efficiency of knockdown was determined by immunoblots. U2OS cells were transfected with siRNA against STIL (5′-AAAUCUUCUGACUCACUGGAUGAGG-3′) (ThermoFisher) or negative control (ThermoFisher catalogue # 4457289).
Immunoblotting
For preparation of cell lysates, cells were washed twice with ice-cold PBS, harvested with a rubber cell scraper, and resuspended in lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and freshly added protease inhibitor cocktail (Roche). After a 30-min incubation, the cell lysates were centrifuged at 1889 × g at 4°C for 10 min. Total protein levels in the lysate supernatants were quantified by Bio-Rad protein assay. Samples with equal amounts of protein were boiled with 4X loading buffer (250 mM Tris pH 6.8, 8% SDS, 40% glycerol, 5% β-mercaptoethanol, 0.2% bromophenol blue [wt/vol]) for 10 min and then separated by 10% SDS–PAGE. Proteins were transferred onto nitrocellulose membranes (Bio-Rad). Membranes were blocked for 30 min at room temperature in PBST (PBS + 0.3% vol/vol Tween-20) containing 5% dry milk and then incubated overnight at 4°C with diluted primary antibodies. Membranes were washed three times with PBST and incubated with horseradish peroxidase–conjugated goat anti-mouse (Jackson Research Laboratories) or donkey anti-rabbit (GE Healthcare) secondary antibodies for 1 h at room temperature, followed by three PBST washes and enhanced chemiluminescence substrate (ThermoFisher). For STIL siRNA, U2OS cells were transfected using an Amaxa Nucleofector II (program X-001). Cell lysates were prepared 48 h posttransfection by lysing cells in 25 mM Tris (pH 7.4), 125 mM NaCl, 1 mM ethylene glycol tetraacetic acid, 0.05% Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol. The Bradford protein assay (Bio-Rad) was used to measure lysate protein concentrations following the manufacturer’s instructions. Laemmli sample buffer was then added, and samples were boiled for 5 min. Equal total protein was loaded for each sample. STIL RNA interference (RNAi) was used determined the specificity of the anti-STIL antibody. Endogenous α-tubulin was used as loading control. Antibodies were used at a concentration of 1–5 μg/ml.
Immunofluorescence
Cells were treated as indicated in the text and then fixed either in prechilled 100% methanol (Figures 1 and 2) at 20°C for 5 min or in 3.7% paraformaldehye (Figure 4) in PBS for 10 min at room temperature. After fixation, cells were rinsed with PBS buffer for three times, and permeabilized in PBS buffer + 0.1% Triton-X for 30 min. Appropriate primary antibodies were diluted in PBS buffer containing 0.1% Triton-X and 0.5% BSA, and samples were incubated for 1 h at room temperature, followed by three washes in PBS to remove unbound primary antibodies. Cells were then incubated with fluorochrome-conjugated secondary antibodies (Molecular Probes) as well as DAPI for 1 h at room temperature and washed three times in PBS. Coverslips were mounted in Fluoromount G Mounting medium (SouthernBiotech). Imaging was performed with a Zeiss LSM 800 confocal microscope (Carl Zeiss) using a Plan-Apochromat 63×/1.4 NA oil objective and appropriate filters as previously described (Xie et al., 2016). Images were acquired by Zen (Carl Zeiss), and processing and analysis (including the mean particle function to calculate vesicles per cell) were performed with ImageJ (National Institutes of Health [NIH], Bethesda, MD). Images were cropped, adjusted for brightness (whole-image adjustment) with minimal manipulation for better presentation, and, when noted, displayed as maximal projections of z-sections. For quantification, three independent experiments were performed, and ImageJ software was used for ratiometric calculations of intensity at the centrosome (or spindle midbody); the number of samples collected is described in the text. For colocalization measurement (Figure 4, D–F), more than 30 cells from three independent confocal microscopy experiments were assessed using ImageJ with Mander’s colocalization coefficient using the JaCoP plug-in.
Live-cell imaging
U2OS cells grown on 35-mm glass-bottom tissue culture dishes (World Precision Instruments) were transfected as indicated in the text. On the day of imaging, the cells were incubated in freshly prepared phenol red–free DMEM (Invitrogen) containing 10% FBS, 2 mM l-glutamine, and 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (ThermoFisher) for at least 15 min. before being transferred to the imaging chamber. For live-cell imaging monitoring the movement of transferrin-containing vesicles, cells were first serum-starved in DMEM containing 2 mM l-glutamine for 1 h and then incubated in the phenol red–free medium along with 25 μg/ml Alexa Fluor 568–transferrin conjugate for 15 min. The transferrin-containing medium was then replaced with fresh phenol red–free medium, and live-cell imaging was initiated. Confocal microscopy imaging was done with a Zeiss LSM 800 microscope (Carl Zeiss) using a Plan-Apochromat 63×/1.4 NA oil objective and appropriate filters. Images in each channel were obtained simultaneously at 512 × 512 pixels with z-slices of 1.5 μm. The images were gathered every 2 s, and z-projections are presented in the figures.
Statistical analysis
Data obtained from ImageJ were exported to Prism 7 (GraphPad). Bar graphs were created representing the mean and SD of the mean from data obtained from three independent experiments. Statistical significance was calculated by nonparametric unpaired two-tailed t test using the Mann-Whitney U-test of unpaired t test data (Figures 2 and 4 and Supplemental Figure 2), by the “N-1” chi-squared test for comparison of proportions (Figure 1 and Supplemental Figure 3) (Campbell, 2007; Richardson, 2011), or by two-way analysis of variance (GraphPad Prism7) (Supplemental Figure 1).
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Qi (Hong Kong University of Science and Technology) for generously providing the GFP-Cep215 plasmid. We acknowledge the following grant support: National Institute of General Medical Sciences (NIGMS) R01GM074876 and P30GM106397 (S.C.) and National Cancer Institute P30 CA23074 and NIH/NIGMS R01GFM110166 and R01GM126035 (G.C.R.).
Abbreviations used:
- DAPI
4′,6-diamidino-2-phenylindole
- FBS
fetal bovine serum
- GST
glutathione- S-transferase
- GFP
green fluorescent protein
- HU
hydroxyurea
- MBP
maltose-binding protein
- PCM
pericentriolar material
- PCNT
pericentrin
- RNAi
RNA interference
- siRNA
small interfering RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-04-0241) on September 6, 2018.
REFERENCES
- Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. (2005). Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol , 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera JA, Kao LR, Hammer RE, Seemann J, Fuchs JL, Megraw TL. (2010). CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell , 913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckson-Castaing V, Moudjou M, Ferguson DJ, Mucklow S, Belkaid Y, Milon G, Crocker PR. (1996). Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J Cell Sci (Pt 1), 179–190. [DOI] [PubMed] [Google Scholar]
- Cai B, Katafiasz D, Horejsi V, Naslavsky N. (2011). Pre-sorting endosomal transport of the GPI-anchored protein, CD59, is regulated by EHD1. Traffic , 102–120. [DOI] [PubMed] [Google Scholar]
- Campbell I. (2007). Chi-squared and Fisher-Irwin tests of two-by-two tables with small sample recommendations. Stat Med , 3661–3675. [DOI] [PubMed] [Google Scholar]
- Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. (2002). A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J , 2557–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. (2002). CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell , 339–350. [DOI] [PubMed] [Google Scholar]
- Conduit PT, Wainman A, Raff JW. (2015). Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol , 611–624. [DOI] [PubMed] [Google Scholar]
- Fong KW, Choi YK, Rattner JB, Qi RZ. (2008). CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol Biol Cell , 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KW, Hau SY, Kho YS, Jia Y, He L, Qi RZ. (2009). Interaction of CDK5RAP2 with EB1 to track growing microtubule tips and to regulate microtubule dynamics. Mol Biol Cell , 3660–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. (1998). C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol , 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giridharan SS, Cai B, Vitale N, Naslavsky N, Caplan S. (2013). Cooperation of MICAL-L1, syndapin2, and phosphatidic acid in tubular recycling endosome biogenesis. Mol Biol Cell , 1776–1790, S1771–S1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokool S, Tattersall D, Seaman MN. (2007). EHD1 interacts with retromer to stabilize SNX1 tubules and facilitate endosome-to-Golgi retrieval. Traffic , 1873–1886. [DOI] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Nigg EA. (2007). Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci , 4321–4331. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. (2005). The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol , 1140–1146. [DOI] [PubMed] [Google Scholar]
- Hehnly H, Doxsey S. (2014). Rab11 endosomes contribute to mitotic spindle organization and orientation. Dev Cell , 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic M, Naslavsky N, Rapaport D, Horowitz M, Caplan S. (2007). EHD1 regulates beta1 integrin endosomal transport: effects on focal adhesions, cell spreading and migration. J Cell Sci , 802–814. [DOI] [PubMed] [Google Scholar]
- Kumar A, Girimaji SC, Duvvari MR, Blanton SH. (2009). Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet , 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Terada Y, Lee KS, Wang CL. (2007). Centrosome replication in hydroxyurea-arrested CHO cells expressing GFP-tagged centrin2. J Cell Sci , 2444–2453. [DOI] [PubMed] [Google Scholar]
- Lee K, Rhee K. (2012). Separase-dependent cleavage of pericentrin B is necessary and sufficient for centriole disengagement during mitosis. Cell Cycle , 2476–2485. [DOI] [PubMed] [Google Scholar]
- Lin SX, Grant B, Hirsh D, Maxfield FR. (2001). Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol , 567–572. [DOI] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Khodjakov A. (2010). Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr Biol , 1277–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang YS, et al. (2015). Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol , 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HT, Poon RY. (2017). Synchronization of HeLa cells. Methods Mol Biol , 189–201. [DOI] [PubMed] [Google Scholar]
- Mardin BR, Schiebel E. (2012). Breaking the ties that bind: new advances in centrosome biology. J Cell Biol , 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Ohsumi K, Iwabuchi M, Kawamata T, Ono Y, Takahashi M. (2012). Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr Biol , 915–921. [DOI] [PubMed] [Google Scholar]
- Moss DK, Bellett G, Carter JM, Liovic M, Keynton J, Prescott AR, Lane EB, Mogensen MM. (2007). Ninein is released from the centrosome and moves bi-directionally along microtubules. J Cell Sci , 3064–3074. [DOI] [PubMed] [Google Scholar]
- Naslavsky N, Boehm M, Backlund PS, Jr., Caplan S. (2004). Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol Biol Cell , 2410–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan JK, Marzio A, Jones MJ, Saraf A, Jallepalli PV, Florens L, Washburn MP, Pagano M. (2015). Degradation of Cep68 and PCNT cleavage mediate Cep215 removal from the PCM to allow centriole separation, disengagement and licensing. Nat Cell Biol , 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser SL, Samant MD, Baxter JE, Morrison CG, Fry AM. (2012). Oscillation of APC/C activity during cell cycle arrest promotes centrosome amplification. J Cell Sci , 5353–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JT. (2011). The analysis of 2 x 2 contingency tables—yet again. Stat Med , 890; author reply 891–892. [DOI] [PubMed] [Google Scholar]
- Simone LC, Caplan S, Naslavsky N. (2013). Role of phosphatidylinositol 4,5-bisphosphate in regulating EHD2 plasma membrane localization. PLoS ONE , e74519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere N, Wagner M, Beishir S, Smith E, Breslin L, Morrison CG, Hochegger H, Kuriyama R. (2011). Centrosome amplification in CHO and DT40 cells by inactivation of cyclin-dependent kinases. Cytoskeleton (Hoboken) , 446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW. (2010). Drosophila Ana2 is a conserved centriole duplication factor. J Cell Biol , 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. (2006a). Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol , 74–78. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. (2006b). Mechanism limiting centrosome duplication to once per cell cycle. Nature , 947–951. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. (2006). Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA , 10660–10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Bahl K, Reinecke JB, Hammond GR, Naslavsky N, Caplan S. (2016). The endocytic recycling compartment maintains cargo segregation acquired upon exit from the sorting endosome. Mol Biol Cell , 108–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeow I, Howard G, Chadwick J, Mendoza-Topaz C, Hansen CG, Nichols BJ, Shvets E. (2017). EHD proteins cooperate to generate caveolar clusters and to maintain caveolae during repeated mechanical stress. Curr Biol , 2951–2962.e2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.