Abstract
Centriole-to-centrosome conversion (CCC) safeguards centriole homeostasis by coupling centriole duplication with segregation, and is essential for stabilization of mature vertebrate centrioles naturally devoid of the geometric scaffold or the cartwheel. Here we identified PPP1R35, a putative regulator of the protein phosphatase PP1, as a novel centriolar protein required for CCC. We found that PPP1R35 is enriched at newborn daughter centrioles in S or G2 phase. In the absence of PPP1R35, centriole assembly initiates normally in S phase, but none of the nascent centrioles can form active centrosomes or recruit CEP295, an essential factor for CCC. Instead, all PPP1R35-null centrioles, although stable during their birth in interphase, become disintegrated after mitosis upon cartwheel removal. Surprisingly, we found that neither the centriolar localization nor the function of PPP1R35 in CCC requires the putative PP1-interacting motif. PPP1R35 is thus acting upstream of CEP295 to induce CCC for proper centriole maintenance.
INTRODUCTION
Centriole homeostasis in animal cycling cells is carefully maintained. In vertebrates, cells begin the cell cycle with two centrioles, each of which is capable of initiating duplication in S phase, generating two mother-daughter centriole pairs. Duplication starts from the formation of the cartwheel upon which other components are assembled. In mitosis, the two pairs of centrioles segregate equally to two daughter cells through association with the spindle poles, restoring the normal copy number of centrioles for the next cell cycle. Centrioles can duplicate and associate with spindle poles only when they have been converted to centrosomes (Wang et al., 2011). The conversion is a process in which newborn centrioles acquire the competence to recruit the pericentriolar material (PCM) and thereby function as the microtubule-organizing center (MTOC) or centrosome (Wang et al., 2011). It starts in early mitosis depending on Plk1 activity, and completes at the end of the cell cycle, giving rise to one old/previously converted and one newly converted centriole that are both MTOC-competent to start the new cell cycle. Only the converted, MTOC-competent centriole can duplicate and then carries the newborn, MTOC-noncompetent daughter centriole to which it is engaged through the segregation process, ensuring centriole homeostasis. Interestingly, concurrent with centriole-to-centrosome conversion (CCC), the cartwheel is removed from newborn centrioles during mitosis, depending also on Plk1. The functional relationship between centrosome formation and cartwheel removal had been largely unknown until the identification of CEP295 as the centriolar factor specifically required for the CCC (Izquierdo et al., 2014; Tsuchiya et al., 2016), but not for cartwheel removal, uncoupling these two events (Izquierdo et al., 2014). In addition, it was reported that CEP295 is required for proper elongation of centrioles (Chang et al., 2016). In Drosophila, both the Polo kinase and the functional homologue of CEP295, Ana1, have been shown to mediate a similar process of CCC (Fu et al., 2016; Novak et al., 2016; Saurya et al., 2016). However, unlike vertebrates, fly centrioles do not lose their cartwheel during the cell cycle.
Human CEP295 is enriched at newborn centrioles in the beginning of centriole duplication in S phase (Izquierdo et al., 2014). Depletion of CEP295 has no effect on old (or mother) centrioles already converted to centrosomes, but it completely abolishes the ability of newborn centrioles to recruit the PCM (e.g., γ-tubulin). In contrast, cartwheel removal from newborn centrioles can still occur normally in the absence of CEP295. Thus, upon acute knockdown of CEP295, cells exiting from mitosis were found to inherit two “cartwheel-less” centrioles: one (the mother) with the PCM and one (the newborn/daughter) without, instead of two cartwheel-less, PCM-associating centrioles normally seen in wild-type G1 cells. These unusual centrioles, devoid of both the cartwheel and PCM, are unstable, becoming disintegrated gradually after mitosis. It has therefore led to the proposal that loss of the cartwheel can be detrimental to centrioles, and that CCC mediated by CEP295 must occur in parallel to stabilize “cartwheel-less” centrioles for duplication. Consistently, in the current study, we identify another centriolar factor named PPP1R35, which acts upstream of CEP295 to promote centriole-to-centrosome conversion, mediating a process fundamentally essential for the stability and homeostasis of centrioles or centrosomes in vertebrate cycling cells.
RESULTS AND DISCUSSION
PPP1R35 is recruited to and enriched at newborn daughter centrioles during S/G2 phase
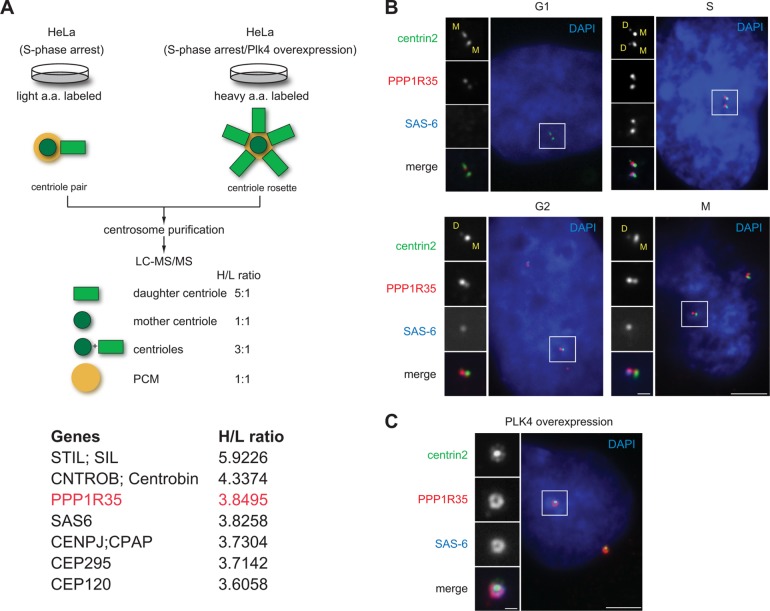
Using the quantitative proteomics SILAC (stable isotope labeling by amino acid in cell culture) screen established previously (Tanos et al., 2013; Wang et al., 2013), we have identified centriolar proteins enriched at newborn daughter centrioles in S phase (Figure 1A). Among them, PPP1R35/C7ORF47, a protein annotated as a putative regulator of the protein phosphatase PP1 (Hendrickx et al., 2009; Fardilha et al., 2011), has not been functionally associated with the centrosome, although it is in the published centrosome proteome (Jakobsen et al., 2011). Using a rabbit polyclonal antibody we generated, which recognizes the N-terminal half of PPP1R35, we found PPP1R35 localizes to all centrioles throughout the cell cycle of diploid, nontransformed retinal pigment epithelial (RPE) cells (Figure 1B). Interestingly, PPP1R35 exhibited preferential localization to newborn daughter centrioles during S, G2, and M phases (Figure 1B), a localization pattern similar to that of CEP295 (Izquierdo et al., 2014). The same localization pattern was also observed in multiple human cell lines that we examined (U2OS, HeLa, and 16HBE; unpublished data). Consistently, when we overexpressed PLK4 to induce the formation of centriole rosettes, PPP1R35 localizes to all the daughter centrioles encircling the mother centrioles, in a localization pattern similar to that of daughter centriole protein SAS-6 known to mark the cartwheel (Figure 1C). The PPP1R35 antibody we generated is specific as the centriolar signals identified diminished upon depletion of PPP1R35 by RNA interference (RNAi) (Figure 2A). Taken together, our findings establish that PPP1R35 is a daughter-enriched centriolar protein.
FIGURE 1:
PPP1R35 is enriched on daughter centrioles. (A) Schematic outlining the SILAC screen used to classify centrosomal proteins (Tanos et al., 2013; Wang et al., 2013). Two populations of differentially labeled centrosomes were purified in equal amounts. One population of centrosomes (labeled with light isotopes) was isolated from cells arrested in S phase, while the other (labeled with heavy isotopes) was from cells arrested in S phase and overexpressing Plk4. The former consisted mainly of one mother and one daughter centriole pair, while the latter was made up of one mother and four to six daughter centrioles complex, or what is commonly known as the centriole rosette. The mixture was subject to a quantitative proteomics analysis SILAC, and proteins that exhibited a high heavy:light (H/L) isotope ratio were considered as daughter centriole protein candidates. A list of candidates identified by the high H/L ratios is shown. Highlighted in red is PPP1R35, which has a H/L ratio similar to that of the daughter centriole protein SAS-6. (B) PPP1R35 localizes to all centrioles throughout the cell cycle, but is more enriched at newborn daughter centrioles in S/G2 phase. Immunofluorescence (IF) images of RPE1 cells at different stages of the cell cycle showing centrin2-GFP (green), PPP1R35 (red), SAS-6 (blue; pseudocolored), and DAPI (blue). Mother (M) and daughter (D) centrioles were marked. Scale bar = 5 μm; 1 μm in inset. (C) Colocalization of PPP1R35 (red) with SAS-6 (blue) at the centriole rosette in PLK4 overexpressing cells. Centrin2-GFP marks centrioles. Scale bar = 5 μm; 1 μm in inset.
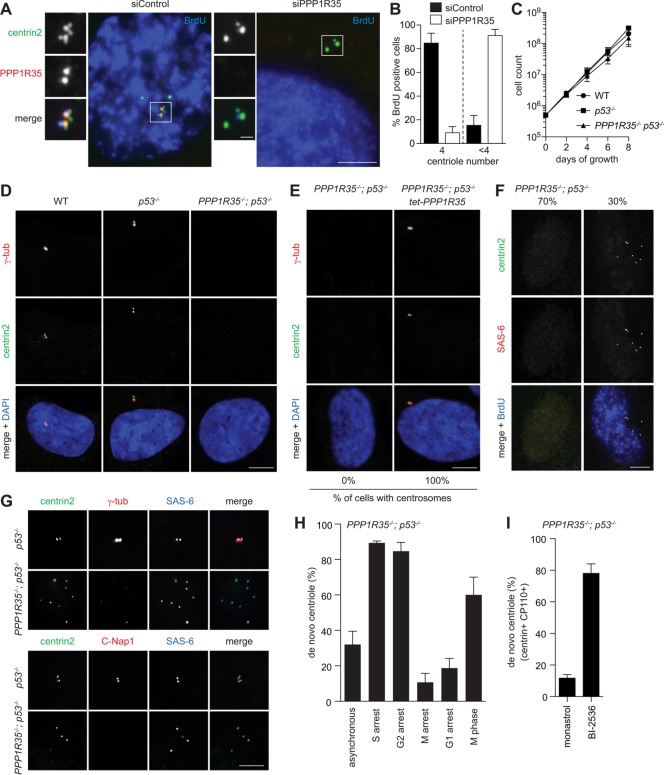
FIGURE 2:
Loss of PPP1R35 causes centriole destabilization during M-G1 transition. (A) PPP1R35 knockdown resulted in centriole number reduction. IF images of RPE1 cells in S phase (BrdU-positive) after PPP1R35 RNAi. Scale bar = 5 μm; 1 μm in inset. (B) Graph showing quantification of phenotypes in A. Data are means ± SD. N > 100, N = 3. (C, D) PPP1R35−/−; p53−/− cells continue to proliferate in the absence of centrosomes. Growth curves (C) and IF images (D) of PPP1R35−/−; p53−/− cells in comparison to wild-type and p53−/− cells. Data are means ± SD. n > 50, N = 3. Scale bar = 5 μm. (E) Centrosome defects in PPP1R35−/−; p53−/− cells are rescued by PPP1R35 expression. PPP1R35−/−; p53−/− cells infected with virus carrying tetracycline-inducible PPP1R35 were examined for the presence of centrosomes. Numbers in the merged image indicate the percentage of cells carrying centrosomes. Scale bar = 5 μm. (F, G) De novo centrioles, all of which carry the cartwheel but not the PCM, are detected in ∼30% of PPP1R35−/−; p53−/− cycling cells. PPP1R35−/−; p53−/− or control (p53−/−) cells growing asynchronously were examined with indicated antibodies. Note that weak centriolar but not strong PCM-associated γ-tubulin is present in de novo centrioles. Scale bar = 5 μm. (H) Centrioles in PPP1R35−/−; p53−/− cells are assembled de novo during S phase, but are disintegrated during M and G1. Graph showing percentage of PPP1R35−/−; p53−/− cells with de novo centrioles at different cell cycle stages. G1-arrest cells were generated by serum starvation for 48 h. S-, G2-, or M-arrest cells were generated by treating cells with aphidicolin, RO-3306, or Eg5 inhibitor for 20 h, respectively. In comparison with M-arrest, normal (nonarrested) M-phase cells in unsynchronized populations were also examined. Data are means ± SD. n > 100, N = 3. (I) PPP1R35−/−; p53−/− cells were allowed to enter mitosis in the presence of the Plk1 inhibitor (BI-2536), or Eg5 inhibitor (monastrol) as a control, and release to and arrest at G1 by Cdk inhibition with roscovitine for 16 h before fixation for examining de novo centrioles. Cells going through this type of manipulation are known to display donut-shaped, multilobed, or multiple small nuclei as described before (Tsou et al., 2009), and were thereby identified for analyses. In Plk1-inhibited cells arrested in G1, de novo centrioles that retained the cartwheel (SAS-6), centrin and CP110 were stably present. Quantifications are shown. Data are means ± SD. n > 100, N = 3.
Loss of PPP1R35 leads to centrosome reduction and centriole disintegration
We found that knockdown of PPP1R35 in asynchronous cycling cells using small interfering RNA (siRNA) resulted in a reduction of centriole number in the majority of cells examined at S phase (Figure 2, A and B), suggesting that PPP1R35 is required for centrosome biogenesis. To confirm and study the impact of PPP1R35 on centrosome biogenesis in detail, we generated a stable RPE1 PPP1R35−/− cell line using CRISPR/Cas9-mediated gene targeting. Because centrosome loss activates a p53-dependent G1 arrest in human cells, the PPP1R35−/− cell line was generated in a p53−/− background. We found that PPP1R35−/−; p53−/− cells proliferate in the absence of centrosomes (Figure 2C), as no γ-tub foci were detected in the cells (Figure 2D). The centrosome loss defect in PPP1R35−/−; p53−/− cells could be rescued by reintroduction of wild-type PPP1R35 into the cells, thus confirming the specificity of the phenotype (Figure 2E).
Despite not having any centrosomes, we could detect multiple centrin foci in ∼30% of PPP1R35−/−; p53−/− cells grown asynchronously (Figure 2F). These centrin foci were found to colocalize with the cartwheel component SAS-6 and other centriolar markers (Figure 2F) but not with the PCM-associated markers (Figure 2G), suggesting that they are not random aggregates but nascent centrioles formed in the absence of PPP1R35. The presence of centriole singlets constantly in a fraction of steady-state, asynchronously growing PPP1R35−/−; p53−/− cells suggests that centrioles are continuously being synthesized de novo and disintegrated during the cell cycle, a phenotype highly similar to that seen in CEP295 knockout cells (Izquierdo et al., 2014). To test this, we synchronized PPP1R35−/−; p53−/− cells in S, G2, M, and G1 phase and monitored the presence of centrioles. We found that de novo centrioles carrying the cartwheel can be detected in the majority of cells arrested in S and G2 phase (Figure 2, F and H), whereas most of the M- or G1-arrested cells do not carry centrioles (Figure 2H). Note that in the unsynchronized PPP1R35−/−; p53−/− population, more than 60% of normal (nonarrested) mitotic cells do carry de novo–formed centrioles, indicating that centriole disintegration starts in M phase and completes after mitotic exit (Figure 2H), the same period of time during which cartwheel removal occurs. To test if centriole disintegration in PPP1R35−/−; p53−/− cells depends on cartwheel removal, cells were allowed to go through mitosis in the absence of Plk1 activity (Figure 2I), a treatment known to block cartwheel removal (Tsou et al., 2009; Izquierdo et al., 2014). Indeed, we found that PPP1R35-null centrioles inhibited of cartwheel removal by Plk1 inactivation can stably exist in G1-arrest cells (Figure 2I), a property similar to that of CEP295-null centrioles. Together, our data demonstrate that centrioles are formed de novo in PPP1R35−/−; p53−/− cells during each S phase, and these centrioles become disintegrated after cartwheel removal in late mitosis and G1. PPP1R35 is hence not required for the initial assembly of centrioles, but it is essential for the stabilization of centrioles losing the cartwheel at the end of mitosis.
PPP1R35 is required for CEP295 recruitment and essential for the conversion of newborn centrioles to centrosomes
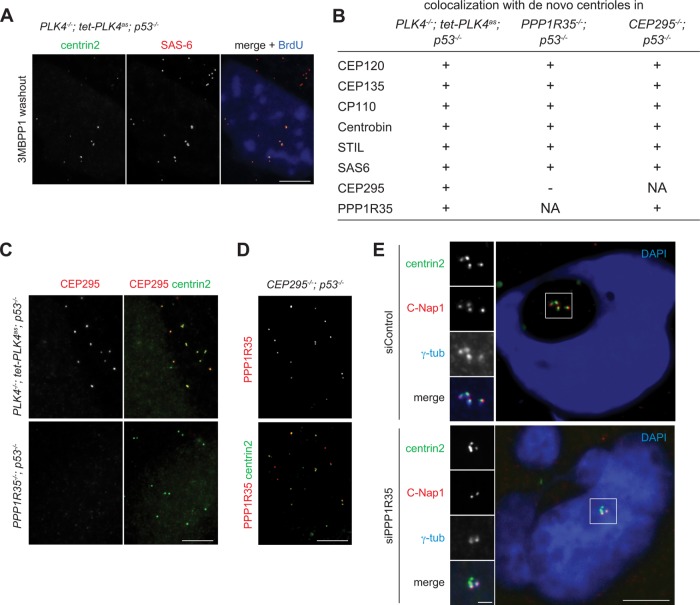
To understand the phenotype seen in PPP1R35−/−; p53−/− cells, we examined the structural composition of the de novo centrioles formed in these cells. For controls, we examined de novo centrioles formed by derepression of the analogue-sensitive PLK4 mutant (PLK4as) in PLK4−/−; p53−/− cells (Figure 3A), which are structurally stable and capable of supporting centrosome duplication. Similar to control cells, de novo centrioles formed in PPP1R35−/−; p53−/− cells also displayed colocalization with multiple centriole markers including CEP120, CEP135, CP110, Centrobin, STIL, and SAS-6 (Figure 3B). However, the daughter-enriched centriole protein CEP295 was absent from the centrioles formed in PPP1R35−/−; p53−/− cells (Figure 3, B and C). Intriguingly, although PPP1R35 is required for CEP295 localization, we found that PPP1R35 can localize to CEP295-null centrioles normally (Figure 3, B and D), indicating that PPP1R35 acts upstream of CEP295.
FIGURE 3:
PPP1R35 is required for CEP295 recruitment and CCC. (A) Derepression of PLK4as generates de novo centrioles. Acentrosomal PLK4−/−; tet-PLK4as; p53−/− cells grown in media containing 3MB-PP1 were washed of 3MBPP1 and arrested in S phase with aphidicolin for 18 h, and processed for immunofluorescence. Scale bar = 5 μm. (B–D) De novo centrioles formed in PPP1R35−/−; p53−/− cells cannot recruit CEP295, but not vice versa. (B) Table showing colocalization of de novo centrioles formed in indicated cell lines with various centriole markers. (C, D) IF images of de novo centrioles in indicated cell lines. Scale bar = 5 μm. (E) PPP1R35 RNAi knockdown cells are defective in CCC. Cells treated with PPP1R35 siRNA were arrested in mitosis with monastrol for 4 h, then released into G1 phase by treatment with CDK1 inhibitor RO-3306 for 4 h, and processed for immunofluorescence. Scale bar = 5 μm; 1 μm in inset.
CEP295 has been shown to be required for the stabilization of newborn centrioles as they exit mitosis. The stabilization is conferred through the acquisition of PCM by the newborn centrioles in a process known as CCC. Once the newborn centriole has been converted to centrosomes, however, CEP295 is no longer needed for the maintenance of the centrosome activity. As PPP1R35 is required for CEP295 localization (Figure 3, B and C), it should have a similar impact on newborn but not mother centrioles. To test this, we used siRNA to knock down PPP1R35, and monitored the short-term effect on both mother and newborn centrioles exiting from mitosis for colocalization with centrosome markers γ-tubulin and C-Nap1. When control mitotic cells, each of which carries four centrioles (two mothers and two newborn daughters), were forced to exit mitosis and enter the G1 phase without undergoing cytokinesis, four centrin foci colocalizing with γ-tubulin and C-Nap1 were detected, indicating that all newborn centrioles underwent CCC (Figure 3E). In contrast, in the absence of PPP1R35, cells exited mitosis with only two out of the four centrin foci carrying the centrosome markers (Figure 3E). These results indicate that PPP1R35 is indeed essential for the conversion of newborn centrioles into centrosomes, consistent with the long-term effect of PPP1R35 depletion seen in our PPP1R35 knockout cells, which completely lack active centrosomes (Figure 2).
Neither the consensus PP1-interacting motif nor the known Plk1 phosphorylation site of PPP1R35 is essential for CCC
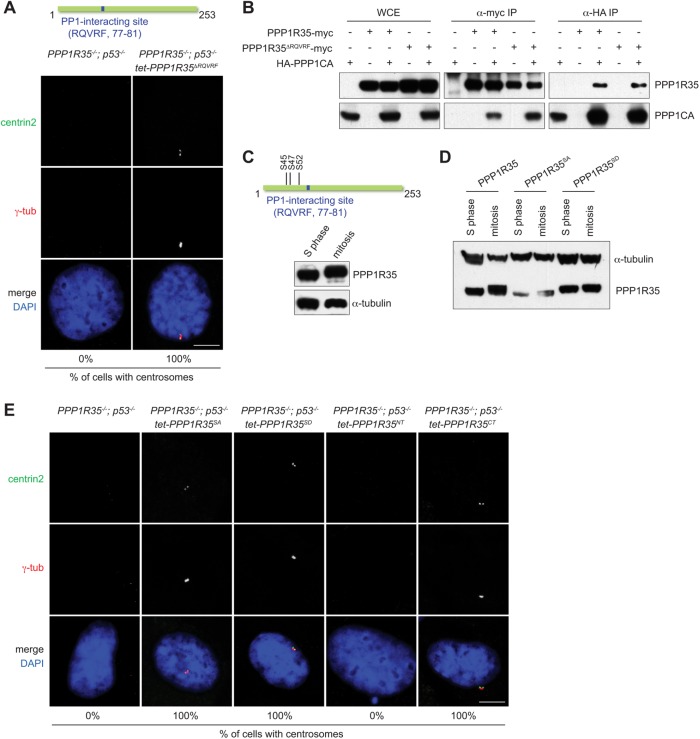
PPP1R35 carries a putative PP1-interacting site (RQVRF; amino acids 77–81) that is conserved in vertebrates, and has been reported to physically associate with the protein phosphatase 1 (PP1) in a large screen for PP1 interactors (Hendrickx et al., 2009; Fardilha et al., 2011). To investigate whether the putative PP1-interacting site of PPP1R35 is required for CCC, we deleted the entire site and examine if the mutant protein (PPP1R35ΔRQVRF) could rescue the CCC defect in PPP1R35−/−; p53−/− cells. We found that centrosomes were formed in all PPP1R35−/−; p53−/− cells upon reintroduction of PPP1R35ΔRQVRF into the knockout cells (Figure 4A), indicating that the RQVRF sequence is not essential for the function of PPP1R35 in CCC. Interestingly, we did not see an interruption to the interaction between PPP1R35ΔRQVRF and PP1 by coimmunoprecipitation experiment (Figure 4B). We then mutated another loosely conserved potential PP1-interacting site (SKRLF; amino acids 157–161; PPP1R35F161A), alone or together with the RQVRF site, but again found that the mutant proteins disrupted neither the function of PPP1R35 in centrosome biogenesis nor its interaction with PP1 (Supplemental Figure S1, A and B). Our data thus suggest that either the interaction of PPP1R35 with PP1 can be nonspecifically created in our and the previous assays (Hendrickx et al., 2009; Fardilha et al., 2011), or there are noncanonical PP1-interacting motifs yet to be identified in PPP1R35.
FIGURE 4:
Neither the PP1-interacting motif nor the known Plk1 phosphorylation site of PPP1R35 is essential for CCC. (A) The PP1-interacting site of PPP1R35 is not essential for CCC. PPP1R35−/−; p53−/− cells stably expressing PPP1R35ΔRQVRF for 2 wk were examined for the presence of centrosomes. Numbers in the merged image indicate the percentage of cells with centrosomes. Scale bar = 5 μm. (B) PPP1R35ΔRQVRF interacts with PP1. HEK293T cells transfected with PPP1R35 and PPP1CA constructs were examined for coimmunoprecipitating proteins. (C) PPP1R35 is phosphorylated during mitosis. A schematic of phophoserine residues on PPP1R35. Immunoblots of PPP1R35 in cells arrested in S phase and mitosis. (D) Phospho-null PPP1R35SA (S45,S47,S52A) and phosphomimetic PPP1R35SD (S45,S47,S52D) mutants of PPP1R35. Immunoblots of phosphorylation mutants of PPP1R35 in cells arrested in S phase and mitosis. (E) Centrosome defects in PPP1R35−/−; p53−/− cells are rescued by PPP1R35 phosphorylation mutants expression. PPP1R35−/−; p53−/− cells stably expressing various indicated PPP1R35 mutants were examined for the presence of centrosomes. PPP1R35NT encodes the first 102 amino acid residues while PPP1R35CT encodes the 103–253 residues. Numbers in the merged image indicate the percentage of cells with centrosomes. Scale bar = 5 μm.
Plk1 is the kinase required for CCC (Wang et al., 2011; Novak et al., 2016). Intriguingly, in a published proteomics screen (Oppermann et al., 2012), PPP1R35 has been identified as a high confident Plk1 substrate during mitosis, with three phosphorylation sites (serine 45, 47, and 52) being mapped in the N-terminal region of the protein (Figure 4C). Indeed, in our gel mobility shift assay, PPP1R35 was found hyperphosphorylated during mitosis, largely depending on these three serine sites (Figure 4, C and D). We thus asked whether PPP1R35 phosphorylation is crucial for its CCC function. We found that expression of either the phosphor-null (PPP1R35SA) or phosphomimetic (PPP1R35SD) mutant protein can fully rescue centrosome formation in PPP1R35−/−; p53−/− cells (Figure 4, D and E). Even more surprisingly, we found that the C-terminal fragment of PPP1R35 alone (103–253 amino acids; PPP1R35CT), which lacks both the identified Plk1 phosphorylation sites and the consensus PP1 interaction motif, could efficiently rescue the centrosome biogenesis defect seen in PPP1R35−/−; p53−/− cells (Figure 4E). In contrast, the N-terminal half of PPP1R35 (1–102 amino acids; PPP1R35NT) was unable to promote or rescue centrosome formation in the knockout cells (Figure 4E). Our data hence demonstrate that the activity of PPP1R35 in driving centrosome biogenesis can be sufficiently supported by the C-terminal half of the protein, independent of the known Plk1 phosphorylation sites and PP1-interacting motif both located in the N-terminus.
We have identified PPP1R35 as a novel daughter centriole enriched protein acting upstream of CEP295 to drive CCC. A recent study showed that PPP1R35 is also involved in centriole elongation (Sydor et al., 2018), similar to CEP295 (Chang et al., 2016). Two previous studies independently identified PPP1R35 as one of more than 70 PP1 phosphatase regulators in human cells (Hendrickx et al., 2009; Fardilha et al., 2011). The interaction of PPP1R35 with PP1 was shown separately by the pull-down assay using bacteria-purified proteins, yeast two-hybrid assay, and coimmunoprecipitation assay (this study). Most of the known PP1 interactors contain the PP1-interacting site, with a consensus sequence as [HKR]-[ACHKMNQRSTV]-V-[CHKNQRST]-[FW]. PPP1R35 can be found in deuterostome animals, but its putative PP1-interacting site (RQVRF) is highly conserved only in vertebrates (100% identical between zebrafish and human). Although our data indicate that the PP1-interacting site of PPP1R35 is not essential for centrosome formation or PP1 association, we anticipate a possibility that it may have an undefined regulatory role associating with other activities of the vertebrate centrosome, perhaps including, for example, the control of ciliogenesis specific for vertebrate cells. More studies are required to test these ideas.
MATERIALS AND METHODS
Cell culture
RPE1 cells were cultured in DME/F-12 (1:1) medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. The RPE1 tetracycline-inducible PLK4as cells (PLK4−/−; tet-PLK4as) were grown under a constant supply of 5 ng/ml doxycycline replaced every 2 d to support centrosome biogenesis. To inhibit PLK4as, 2 μM of 3MBPP1 was added to the media. To reactivate PLK4as, cells were washed five times with fresh media to remove 3MBPP1. BrdU was added to culture medium at 30 μM for 30 min before fixation to label S-phase cells. Aphidicolin (2 μg/ml) was used to arrest cells in S phase. Cdk1 inhibitor RO-3306 (10 μM) was used to arrest cells at G2/M boundary. For M phase arrest, cells were treated with 200 ng/ml nocodazole. To arrest cells in G1, cells were washed three times with serum-free media and grown in serum-free media for 2 d.
Cell lines and plasmid constructs
Generation of a stable clonal PPP1R35−/−; p53−/− cell line is described here. For rescue experiments, a clonal PPP1R35−/−; p53−/− cell line stably carrying various constructs expressing PPP1R35WT, PPP1R35ΔRQVRF, PPP1R35SA (S45,S47,S52A), PPP1R35SD (S45,S47,S52D), PPP1R35NT (1-102aa), PPP1R35CT (103-253aa), PPP1R35F161A, and PPP1R35doublePP1 (R76,R77,F81,F161A) from the tetracycline-inducible promoter were made through in vivo gene delivery using the lentiviral vector pLVX-Tight-Puro vector (Clonetech). The RPE1 tetracycline-inducible PLK4as cell line (PLK4−/−; tet-PLK4as) was generated in our lab (Kim et al., 2016). PPP1R35 cDNA construct was obtained from Open Biosystems. PPP1R35ΔRQVRF, PPP1R35SA (S45,S47,S52A), PPP1R35SD (S45,S47,S52D), PPP1R35NT (1-102aa), PPP1R35CT (103-253aa), PPP1R35F161A, and PPP1R35doublePP1 (R76,R77,F81,F161A) constructs were created with site-directed mutagenesis (Stratagene), and were used for subcloning into pcDNA3-FLAG-HA and pLVX-Tight-Puro vector. PPP1CA was amplified from a cDNA library from HeLa cells made in our lab, and was cloned into pcDNA3-FLAG-HA vector.
CRISPR-mediated gene targeting
RNA-guided targeting of PPP1R35 in human cells was achieved through coexpression of the Cas9 protein with guide RNAs (gRNAs) using reagents as described previously (Mali et al., 2013), which are available from Addgene (www.addgene.org/crispr/church/). Three gRNAs were used together to knock out PPP1R35 in RPE1 cells. Sequences of PPP1R35 gRNAs used are as follows: (5′-GAGTCAGAGCTGAAGTCGG-3′), (5′-GCGGACGGGGAAGAAGCCG-3′), and (5′-GCCTTGGGCCTGGAGCTGC-3′). All gRNAs were cloned into the gRNA cloning vector (Addgene plasmid #41824) via the Gibson assembly method (New England Biolabs) as described previously (Mali et al., 2013). Cas9 plasmid (5 μg; Addgene plasmid #41815) and gRNA (5 μg) were nucleofected according to manufacturer’s instructions (Lonza). Cells were examined for the loss of proteins and centrosomes 7 d after nucleofection.
RNAi and stable cell lines
Transient transfection of siRNA oligos was performed using RNAiMAX (Life Technologies). The PPP1R35 siRNA oligo used was Silencer Select s195859 (ThermoFisher Scientific). For a complete knockdown of PPP1R35, three sequential siRNA oligo transfections were performed every 2 d and cells were harvested at day 6. Stable clones of RPE-1 cells inducibly expressing the various PPP1R35 constructs from the tetracycline-inducible promoter were obtained through in vivo gene delivery using the lentiviral vector pLVX-Tight-Puro vector (Clonetech).
Antibodies
Antibodies used in this study are listed with the information on working dilution and source in parentheses: anti-centrin2 (mouse, 1:1000, 04-1624; Millipore), anti-γ-tub (mouse, 1:500, sc-51715; Santa Cruz Biotechology), anti-BrdU (rat, 1:500, MCA2060T; AbD Serotec), anti-SAS-6 (mouse, 1:200, sc-81431; Santa Cruz Biotechnolocy), anti-HA (mouse, 1:1000, MMSH101P; Covance; rat, 1:1000, 11867423001; Roche Diagnostic), anti-c-myc (mouse, 1;1000, sc-40; Santa Cruz Biotechnology), anti-STIL (rabbit, 1:200, A302-441A; Bethyl Laboratories), anti-CP110 (rabbit, 1:200, 12780-1-AP; Proteintech), anti-CEP135 (rabbit, 1:200; kind gift from Erich Nigg at University of Basel), anti-CEP120 (rabbit, 1:5; kind gift from Moe R. Mahjoub at Washington University), anti-α-tubulin (mouse, 1:10,000, T6199; Sigma), anti-centrobin (rabbit, 1:5; kind gift from K. Rhee at Seoul National University), anti-C-Nap1 (rabbit, 1:200; produced against the human C-Nap1 as previously described [Tsou and Stearns, 2006]), anti-CEP295 (rabbit, 1:1000, ab122490; Abcam). PPP1R35 polyclonal antibody was generated in rabbits with full-length PPP1R35 SDS–PAGE gel slice. The rabbit serum was subjected to affinity purification using the N-terminal fragment of PPP1R35 (1-127aa). Secondary antibodies Alexa-Fluor 488, 594, and 680 were from Molecular Probes.
Immunofluorescence and microscopy
Cells were washed once in phosphate-buffered saline (PBS) and then fixed in ice-cold methanol at −20°C for 10 min. For immunostaining using the rabbit polyclonal anti-PPP1R35 antibody, cells were washed once in PBS then fixed in 2% paraformaldehyde for 10 min at room temperature. Slides were blocked with 3% bovine serum albumin (BSA; wt/vol) with 0.1% Triton X-100 in PBS before incubating with primary antibodies. For BrdU staining, cells were treated with 2 N HCl at room temperature for 30 min followed by rinsing in PBS before anti-BrdU incubation. DNA was visualized using 4′,6-diamidino-2-phenylindole (DAPI). Fluorescent images were acquired on an upright microscope (Axio imager; Carl Zeiss) equipped with 100× oil objectives, NA of 1.4, and a camera (ORCA ER; Hamamatsu Photonics). Captured images were processed with Axiovision (Carl Zeiss) and Photoshop CS5 (Adobe).
Immunoprecipitation
HEK293T cells were transfected with plasmids using calcium phosphate transfection. Cells were harvested 2 d after in NP-40 lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 10% glycerol) supplemented with Complete protease inhibitors (Roche) at 4°C for 15 min. Lysates were cleared by centrifugation and were subjected to preclearing with Dynabeads Protein G (Invitrogen) for 1 h. Cleared lysates were supplemented with 1% BSA to reduce nonspecific binding to Dynabeads in the subsequent steps. Incubation with antibodies was performed at 4°C for 2 h. Dynabeads Protein G (preincubated in 1% BSA) was subsequently added for an additional hour. Beads were washed in NP-40 lysis buffer and boiled for 3 min in SDS lysis buffer. The extracts were resolved by SDS–PAGE.
Growth curve analysis
Cells were seeded at 500,000 cells per 10-cm plates at day 0. Cells were trypsinized for cell counting using a hemocytometer every 2 d for 8 d and reseeded with appropriate dilution for subsequent counting.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant no. GM-088253 and American Cancer Society Grant no. RSG-14-153-01-CCG to M.-F.B.T. M.-F.B.T. was also supported by the Geoffrey Beene Cancer Research Center and the Functional Genomics initiative.
Abbreviations used:
- CCC
centriole-to-centrosome conversion
- IF
immunofluorescence
- MTOC
microtubule-organizing center
- PCM
pericentriolar material
- RNAi
RNA interference
- RPE
retinal pigment epithelial
- siRNA
small interfering RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0525) on September 19, 2018.
REFERENCES
- Chang CW, Hsu WB, Tsai JJ, Tang CJ, Tang TK. (2016). CEP295 interacts with microtubules and is required for centriole elongation. J Cell Sci , 2501–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardilha M, Esteves SL, Korrodi-Gregorio L, Vintem AP, Domingues SC, Rebelo S, Morrice N, Cohen PT, da Cruz e Silva OA, da Cruz e Silva EF. (2011). Identification of the human testis protein phosphatase 1 interactome. Biochem Pharmacol , 1403–1415. [DOI] [PubMed] [Google Scholar]
- Fu J, Lipinszki Z, Rangone H, Min M, Mykura C, Chao-Chu J, Schneider S, Dzhindzhev NS, Gottardo M, Riparbelli MG, et al. (2016). Conserved molecular interactions in centriole-to-centrosome conversion. Nat Cell Biol , 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. (2009). Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol , 365––371.. [DOI] [PubMed] [Google Scholar]
- Izquierdo D, Wang WJ, Uryu K, Tsou MF. (2014). Stabilization of cartwheel-less centrioles for duplication requires CEP295-mediated centriole-to-centrosome conversion. Cell Rep , 957––965.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, et al. (2011). Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J , 1520–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, O’Rourke BP, Soni RK, Jallepalli PV, Hendrickson RC, Tsou MB. (2016). Promotion and suppression of centriole duplication are catalytically coupled through PLK4 to ensure centriole homeostasis. Cell Rep , 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. (2013). RNA-guided human genome engineering via Cas9. Science , 823––826.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak ZA, Wainman A, Gartenmann L, Raff JW. (2016). Cdk1 phosphorylates Drosophila Sas-4 to recruit Polo to daughter centrioles and convert them to centrosomes. Dev Cell , 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann FS, Grundner-Culemann K, Kumar C, Gruss OJ, Jallepalli PV, Daub H. (2012). Combination of chemical genetics and phosphoproteomics for kinase signaling analysis enables confident identification of cellular downstream targets. Mol Cell Proteomics , O111 012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurya S, Roque H, Novak ZA, Wainman A, Aydogan MG, Volanakis A, Sieber B, Pinto DM, Raff JW. (2016). Drosophila Ana1 is required for centrosome assembly and centriole elongation. J Cell Sci , 2514–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydor AM, Coyaud E, Rovelli C, Laurent E, Liu H, Raught B, Mennella V. (2018). PPP1R35 is a novel centrosomal protein that regulates centriole length in concert with the microcephaly protein RTTN. eLife , e37846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MF. (2013). Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev , 163––168.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. (2006). Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol , 74–78. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. (2009). Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell , 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Yoshiba S, Gupta A, Watanabe K, Kitagawa D. (2016). Cep295 is a conserved scaffold protein required for generation of a bona fide mother centriole. Nat Commun , 12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Soni RK, Uryu K, Tsou MF. (2011). The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J Cell Biol , 727–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Tay HG, Soni R, Perumal GS, Goll MG, Macaluso FP, Asara JM, Amack JD, Tsou MF. (2013). CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nat Cell Biol , 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.