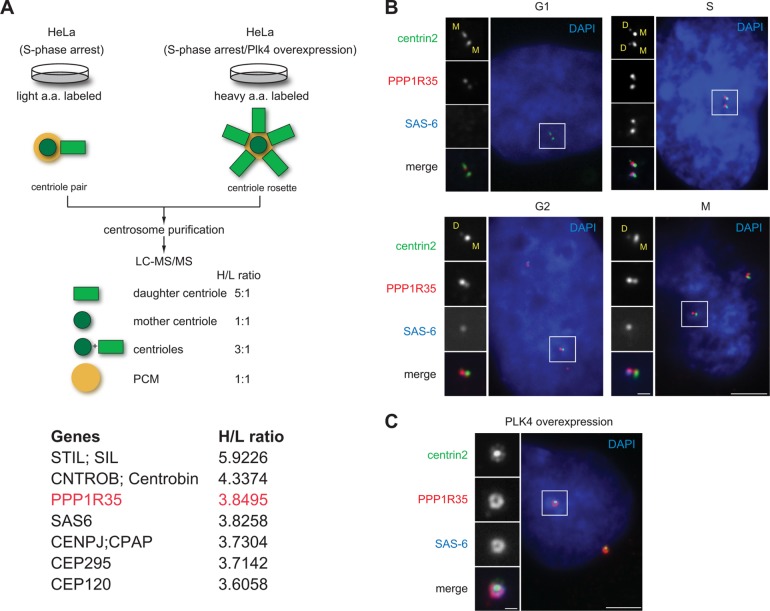
FIGURE 1:
PPP1R35 is enriched on daughter centrioles. (A) Schematic outlining the SILAC screen used to classify centrosomal proteins (Tanos et al., 2013; Wang et al., 2013). Two populations of differentially labeled centrosomes were purified in equal amounts. One population of centrosomes (labeled with light isotopes) was isolated from cells arrested in S phase, while the other (labeled with heavy isotopes) was from cells arrested in S phase and overexpressing Plk4. The former consisted mainly of one mother and one daughter centriole pair, while the latter was made up of one mother and four to six daughter centrioles complex, or what is commonly known as the centriole rosette. The mixture was subject to a quantitative proteomics analysis SILAC, and proteins that exhibited a high heavy:light (H/L) isotope ratio were considered as daughter centriole protein candidates. A list of candidates identified by the high H/L ratios is shown. Highlighted in red is PPP1R35, which has a H/L ratio similar to that of the daughter centriole protein SAS-6. (B) PPP1R35 localizes to all centrioles throughout the cell cycle, but is more enriched at newborn daughter centrioles in S/G2 phase. Immunofluorescence (IF) images of RPE1 cells at different stages of the cell cycle showing centrin2-GFP (green), PPP1R35 (red), SAS-6 (blue; pseudocolored), and DAPI (blue). Mother (M) and daughter (D) centrioles were marked. Scale bar = 5 μm; 1 μm in inset. (C) Colocalization of PPP1R35 (red) with SAS-6 (blue) at the centriole rosette in PLK4 overexpressing cells. Centrin2-GFP marks centrioles. Scale bar = 5 μm; 1 μm in inset.