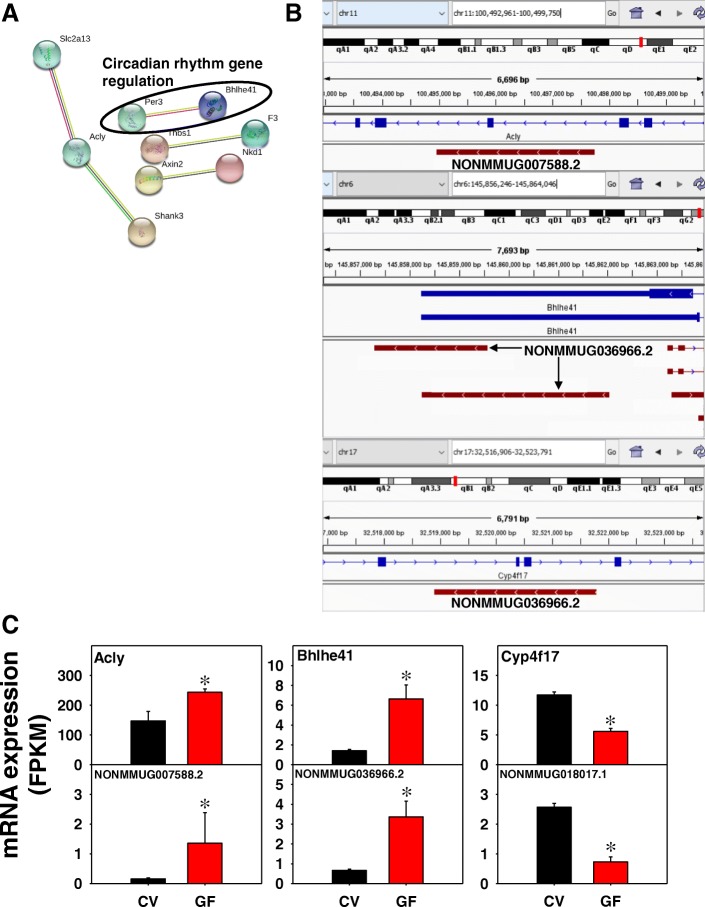
Fig. 12.
Pathway analysis (a), as well as genomic location (b) and gene expression (c) of lncRNA-PCG pairs that were differentially regulated by lack of gut microbiota in WAT of CV and GF mice. (a) The PCGs paired with lncRNAs between CV and GF mice in BAT were subjected to STRING analysis using the default settings. The connected nodes are shown. (b) and (c) The cytosolic acetyl-CoA synthesis enzyme ATP citrate lyase (Acly), the circadian rhythm regulatory gene basic helix-loop-helix family, member e41 (Bhlhe41), and the phase-I oxidation enzyme Cyp4f17 are shown. Expression of lncRNAs and paired PCGs were plotted using mean FPKM ± S.E., and asterisks (*) indicate statistically significant differences between enterotypes of mice (FDR-BH < 0.05)