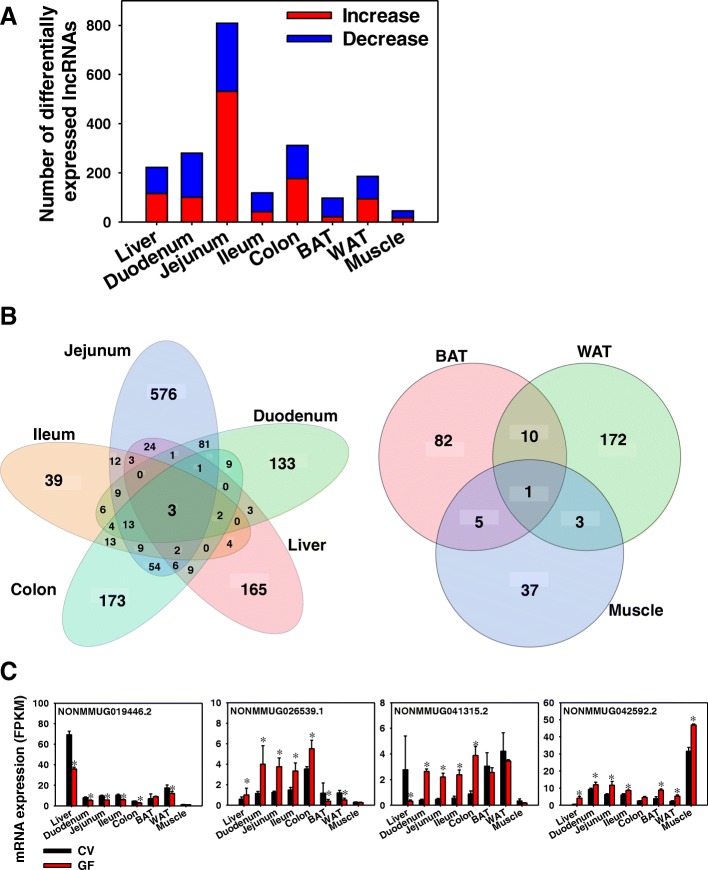
Fig. 2.
Distribution of differentially expressed lncRNAs in eight organs between CV and GF mice. (a) Bar chart showing the number of expressed lncRNAs (average FPKM > 1 in either CV or GF conditions) that were differentially regulated by lack of gut microbiota in liver, duodenum, jejunum, ileum, colon, BAT, WAT, and muscle (FDR-BH < 0.05). (b) Venn diagram showing the number of differentially expressed lncRNAs by lack of gut microbiota in each organ. Liver and various sections of intestine that are proximal to gut micrboiome were grouped together; whereas the energy metabolism related organs (BAT, WAT, and muscle) that are distal to gut microbiota were grouped together. Venn diagram was generated using JMP Genomics