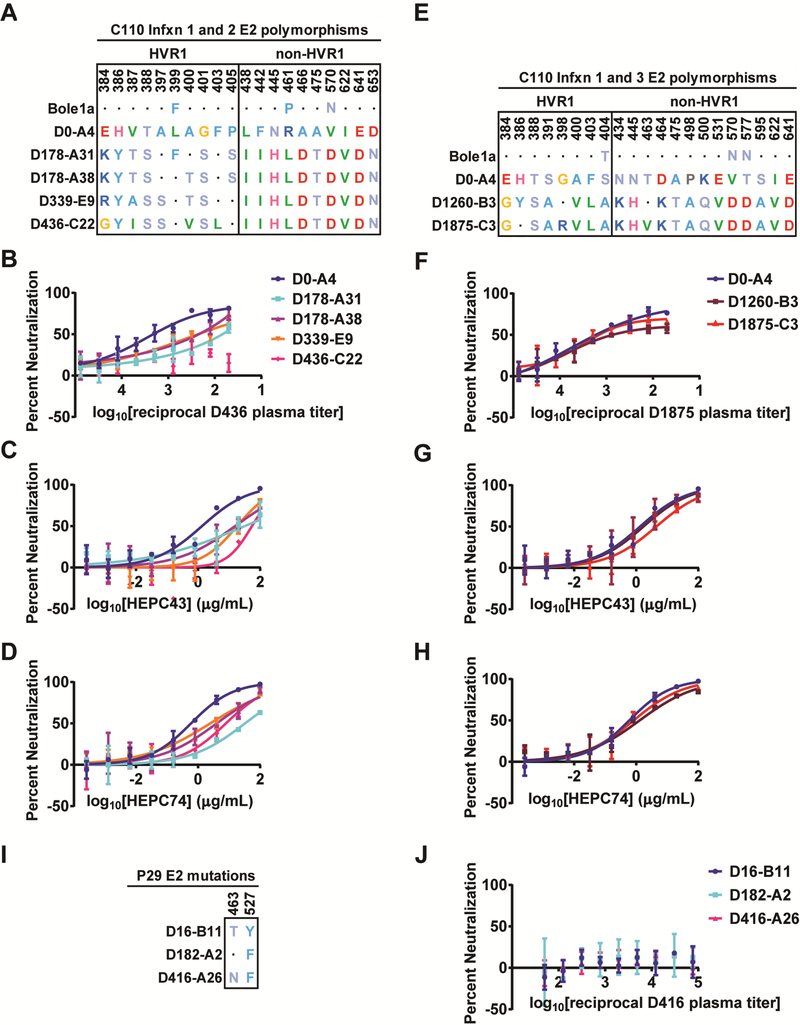
Figure 4. Naturally selected substitutions in C110 E2 confer partial resistance to autologous plasma antibodies and autologous bNAbs.
(A) E2 polymorphisms where the D178-A31 (Infxn 2 T/F) sequence differs from both D0-A4 (Infxn 1) and Bole1a (an inferred ancestral reference sequence), and substitutions arising over the course of Infxn 2. Homology to the D0-A4 sequence is indicated with a dot. (B-D) Neutralization of HCVpp expressing longitudinal C110 E1E2 strains from Infxn 1 and 2 by D436 C110 plasma (B), HEPC43 (C), or HEPC74 (D). (E) E2 polymorphisms where the D1260-B3 (Infxn 3) sequence differs from both D0-A4 (Infxn 1) and Bole1a, and substitutions arising over the course of Infxn 3. Homology to the D0-A4 sequence is indicated with a dot. (F-H) Neutralization of HCVpp expressing longitudinal C110 E1E2 strains from Infxns 1 and 3 by D1875 C110 plasma (F), HEPC43 (G), or HEPC74 (H). (I) Substitutions arising in longitudinal P29 E1E2 clones. (J) Neutralization of HCVpp expressing longitudinal P29 E1E2 strains by autologous D416 plasma. All assays were performed in duplicate, and values are means +/− SD. (See Fig S5 for additional independent experiments.)