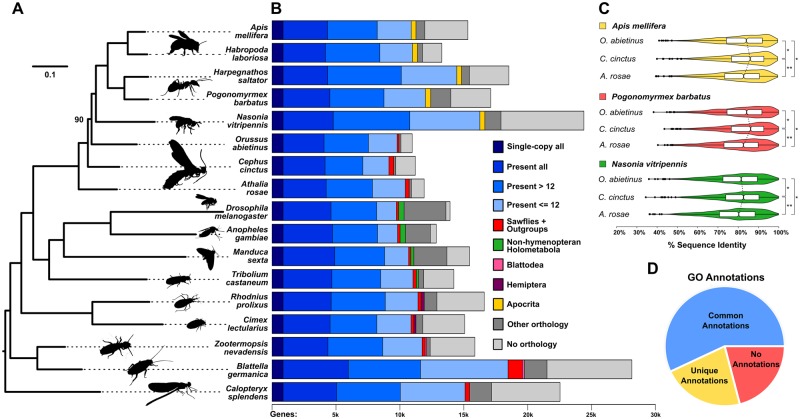
Fig. 2.
—Phylogenomics, orthology, sequence identity, and functional annotation of Cephus cinctus genes. (A) Maximum likelihood molecular phylogeny estimated from the aligned protein sequences of 852 single-copy orthologs from 17 selected insect species, including three sawflies, Athalia rosae, C. cinctus, and Orussus abietinus, with the banded demoiselle, Calopteryx splendens (Odonata), as the outgroup. Branch lengths represent substitutions per site, all nodes except for the placement of O. abietinus (labeled) achieved 100% bootstrap support. (B) Total gene counts per species partitioned into categories from single-copy orthologs in all 17 species, present (i.e., allowing gene duplications) in all or most (>12) or with patchy species distributions (≤12), to lineage-restricted orthologs (sawflies and outgroups, non-hymenopteran Holometabola, Blattodea, Hemiptera, or Apocrita), genes with orthologs from other sequenced insect genomes, and those with currently no identifiable orthologs. (C) Distributions of percent amino acid identities of 1,035 single-copy orthologs from each of the three sawflies with Apis mellifera, Pogonomyrmex barbatus, and Nasonia vitripennis. Violin plots show the smoothed densities of each distribution, with boxplots indicating medians and upper and lower quartiles. Asterisks indicate statistically significant differences with P values <1e-108 (**) and <1e-39 (*). (D) Functional inferences for C. cinctus genes from Gene Ontology (GO) annotations of orthologs from A. mellifera, Anopheles gambiae, Drosophila melanogaster, or Tribolium castaneum. Of the 7,595 C. cinctus genes with orthologs from any of these four species, 57% were in orthologous groups with GO terms shared among at least two of the four species (common annotations), 22% with GO terms from only one species (unique annotations), and 21% where no GO terms were associated with any of the orthologs (no annotations).