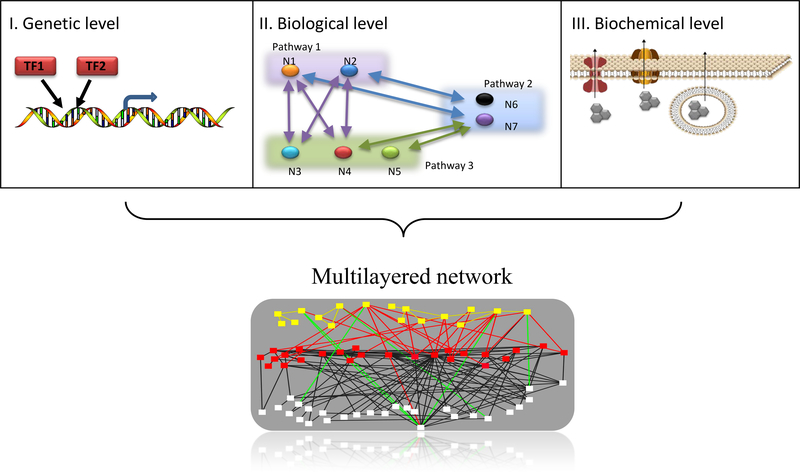
Figure 1. Mechanisms of redundancy.
Redundancy, in general, describes a scenario in which different elements may potentially act in the same biological or dynamic manner, where the inhibition of one of these elements has no significant effect on the global biological outcome or on the system’s dynamic behavior. The redundant elements may refer to the fundamental variables, paths or subsystems that construct the system. Redundancy in biology is a problem known and considered for many years; however, it is essentially unsolved. Unfortunately, this familiar problem is still at the core of many cancer-related open questions that must be addressed. Redundancy occurs at many levels, such as at the genetic level (e.g., two TFs bind to a shared promoter), at the level of biochemical function (e.g., crosstalk among pathways composed of several redundant interactions and variables), and at the level of biological function (e.g. three different ways to efflux a drug from a cell). These different levels have not previously been well-enough defined to be addressed, mainly due to the lack of data and an inadequate understanding of how cancer cells compensate in each of these levels. Today, with advances in technology that enable the collection of different types of high-throughput data sets from a patient, and with new computational methods that have been designed to integrate ‘big-data’ sets into a meaningful representation of a patient’s information, we have a good opportunity to make a renewed effort to find creative ways to solve this problem.