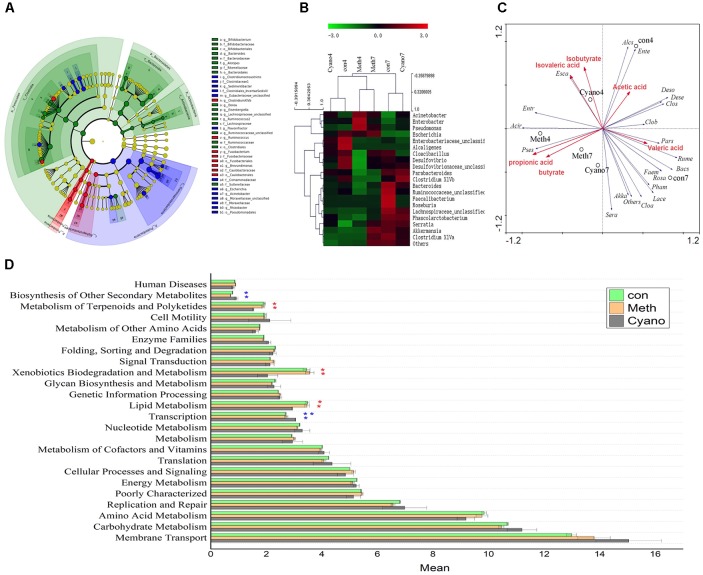
FIGURE 3.
(A) Analysis of taxonomic abundances using LEfSe indicates that multiple taxa are differentially enriched in the three groups (The red color indicate Cyano group, the green color indicate con group, the blue color indicate con group). (B) Heatmap and hierarchical clustering (Euclidean distance) on the taxonomic genus level. (C) Canonical correspondence analysis (“Acir” means Acinetobacter, “Bacs” means Bacteroides, “Ente” means Enterobacteriaceae, “Rume” means Ruminococcaceae, “Faem” means Faecalibacterium, “Entr” means Enterobacter, “Cloa” means Clostridium XlVa, “Pars” means Parabacteroides, “Rosa” means Roseburia, “Lace” means Lachnospiraceae, “Clos” means Cloacibacillus, “Pham” means Phascolarctobacterium, “Alcs” means Alcaligenes, “Pses” means Pseudomonas, “Clob” means Clostridium XlVb, “Deso” means Desulfovibrio, “Esca” means Escherichia, “Sera” means Serratia, “Dese” means Desulfovibrionaceae, “Akka” means Akkermansia) (D) Comparisons of the predominant gene pathways of the microbiota in three groups.