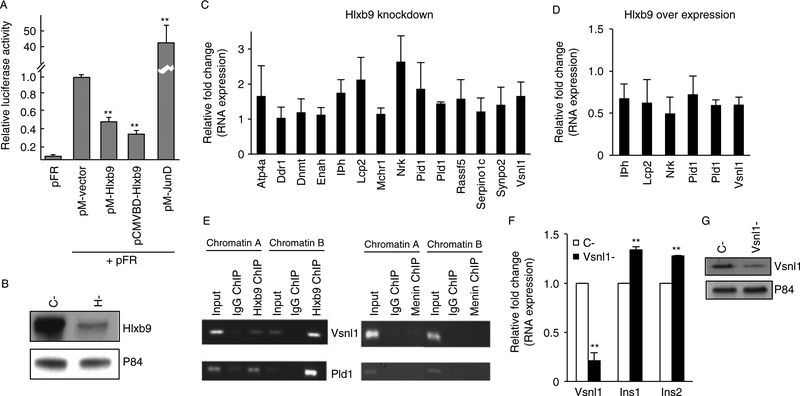
Figure 3.
Hlxb9 target genes in MIN6 cells. (A) Hlxb9 represses transcription. Two different Gal4-DBD fused Hlxb9 constructs (pM-HB9 and pCMVDBDHB9) were assessed for transcriptional activity on a Gal4-UAS-luciferase reporter (pFR). Hlxb9 repressed transcription (luciferase expression) by1.54-fold (pM-HB9) or 2.78-fold (pCMVDBD-HB9). pM-JunD served as a positive control for activating transcription. **Significant effect (P<0.01).(B) Hlxb9 knockdown for gene expression microarray analysis. Three independent transient transfections were performed with control siRNA (C-) or Hlxb9 siRNA (H-). A representative western blot from one such transfection is shown. (C) Validation of Hlxb9 target genes identified by microarray analysis. RNA samples processed for microarray analysis (n=3) and one additional RNA sample of C- and H- were used for QRT-PCR. Of 16 target genes, Palld and Krt4 did not yield a PCR product. QRT-PCR data were plotted as fold change in expression over C- RNA samples (equal to 1). Eleven out of the 12 target genes were significantly upregulated (P<0.05).(D) Hlxb9 target genes upon Hlxb9 overexpression. RNA samples (n=3) from cells transfected with mychis-vector (mh-Vector) or mychis-Hlxb9 (mh-Hlxb9) were assessed for the expression of six target genes, which showed highest significant fold decrease in H-. Data were plotted as fold change in expression over mh-Vector-transfected RNA sample (equals to 1). All six target genes were significantly downregulated (P<0.05). (E) Hlxb9 occupies the promoters of Vsnl1 and Pld1. Primer pairs corresponding to evolutionarily conserved promoter regions of Vsnl1 and Pld1 were used for PCR from anti-Hlxb9 (left panel) or anti-menin (right panel) ChIPs. ChIP with mouse or rabbit IgG served as negative controls for Hlxb9 ChIP and menin ChIP respectively. Shown are ethidium bromide-stained agarose gels of the indicated PCR products from two different ChIP experiments (chromatin A and chromatin B). Input corresponds to 1/50th of chromatin that was used for ChIP. (F and G) Vsnl1 knockdown increases insulin mRNA in MIN6 cells. RNA samples from control siRNA (C-) or Vsnl1 siRNA (Vsnl1-) transfected cells (n=3) were analyzed by QRT-PCR using primer pairs specific for Vsnl1, insulin 1 (Ins1), and insulin 2 (Ins2). Data are shown as fold change in expression compared to control siRNA transfectedcells (equal to 1), **P<0.01. Vsnl1 knockdown was confirmed by western blot (G).