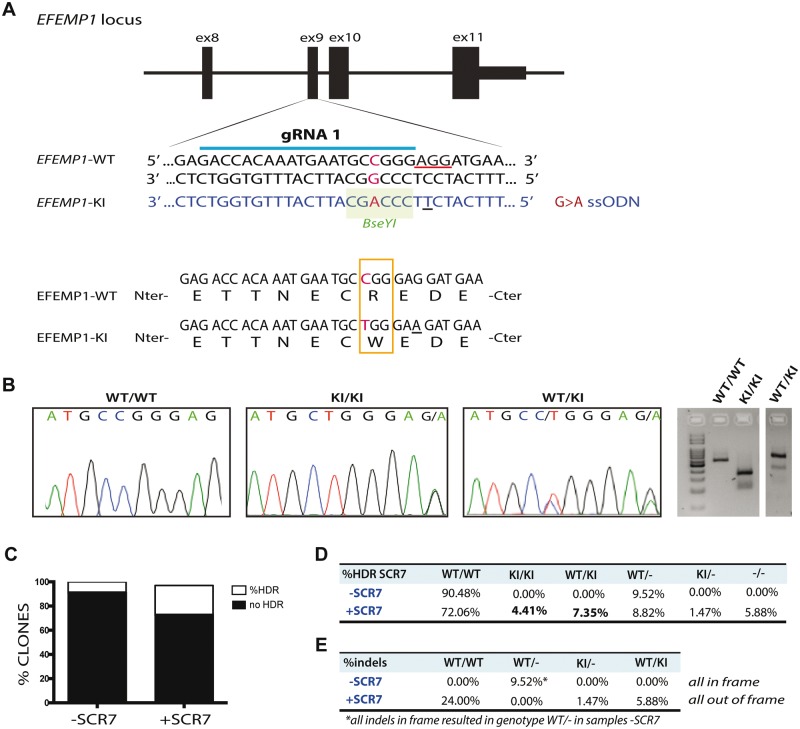
Figure 1.
Knock-in the mutation p.R345W in the EFEMP1 gene via CRISPR–Cas9. (A) Design of guide RNA (gRNA) and single strand oligonucleotide donor (ssODN) to knock-in the mutation c.1033C>T in the exon 9 of the EFEMP1 gene in ARPE-19 cells via CRISPR–Cas9. PAM sequence underlined in red. Change G>A in the ssODN creates a new restriction site for the BseYI enzyme (highlighted in green). (B) Sanger sequence and agarose gel of successfully edited and isolated clones with genotype WT/WT (wild-type), KI/KI (two copies of the mutation, one per allele) and WT/KI (only one allele has the mutation). (C) Percentage of clones that had the mutation, comparing untreated (−SCR7) and treated (+SCR7) with the DNA ligase IV inhibitor SCR7. (D) Quantification of edited clones according to their genotype: WT/WT: both alleles wild-type; KI/KI: both alleles mutant; WT/KI: one allele mutant and one WT; WT/−: one allele wild-type and one allele with indels that result in null allele; KI/−: one allele with the mutation and one allele with indels that result in null allele; −/−: both alleles have indels that result in null alleles. (E) Percentage of clones treated with or without SCR7 which alleles had indels in frame or out of frame.