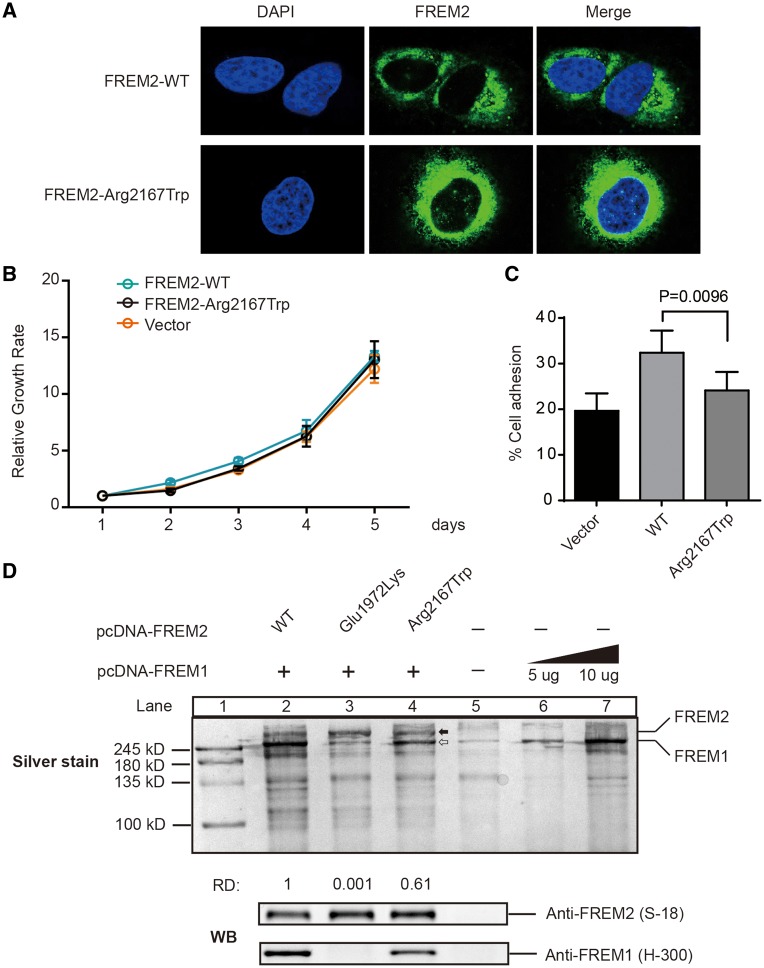
Figure 3.
Function of FREM2 mutation p.Arg2167Trp. (A) Cellular localization of FREM2–WT and FREM2–Arg2167Trp mutant in HEK293T treated with the siRNA and reintroduced it by transfecting siRNA-resistant plasmids: pFREM2–WT or pFREM2–Arg2167Trp. Cells were stained with anti-FREM2 antibody (S-12, Santa Cruz). Nuclei was stained by DAPI (Blue). (B) Growth curve analysis of HEK293T cells transfected with either FREM2–WT or FREM2–Arg2167Trp. Control was done in cells transfected with pcDNA3.1. Error bars represent SEM of six independent experiments. (C) Fibronectin adhesion assay of HEK293T treated with WT or mutant FREM2 as described in Materials and Methods. Error bars represent SEM of six independent experiments. (D) Silver stained and immunoblotted SDS-PAGE gel. Lane 1 is ladder; Lanes 2–5 are immunoprecipitated FREM2 using monoclonal antibody anti-FREM2 (F-1, Santa Cruz); Lanes 6 and 7 immunoprecipitated FREM1 using anti-FREM1 (H-300), solid arrow indicates FREM1 and open arrow indicates FREM2, which is confirmed by immunoblotting with anti-FREM1 (H-300, Santa Cruz) and FREM2 (S-18, Santa Cruz) antibodies (bottom panel). Quantification of the band's density was performed using ImageJ. The capacity of binding of FREM1 was calculate using formulas: . The capacity of binding of FREM1 in Lane 2 was set as 1, and the number under Lanes 3 and 4 indicates the relative capacity of the mutant FREM2 as compared with WT (Lane 2). RD, relative capacity of coimmunoprecipitated FREM1.