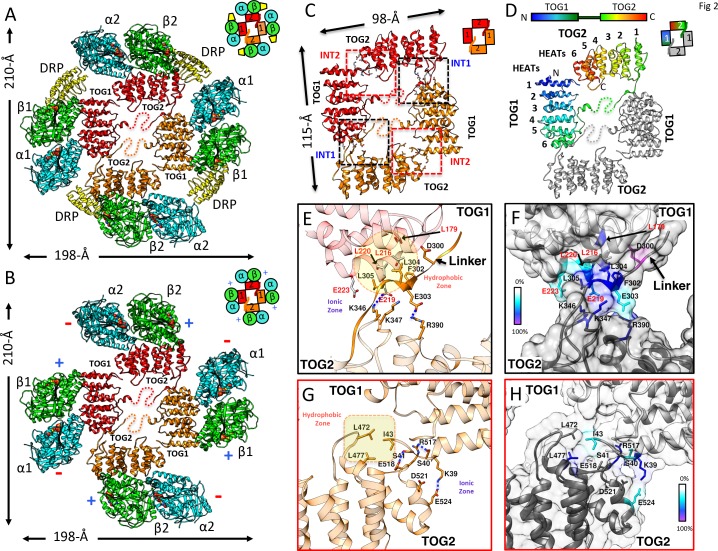
Figure 2. X-ray structures reveal αβ-tubulins bound in a wheel-like organization around a pseudo-dimeric TOG square complex.
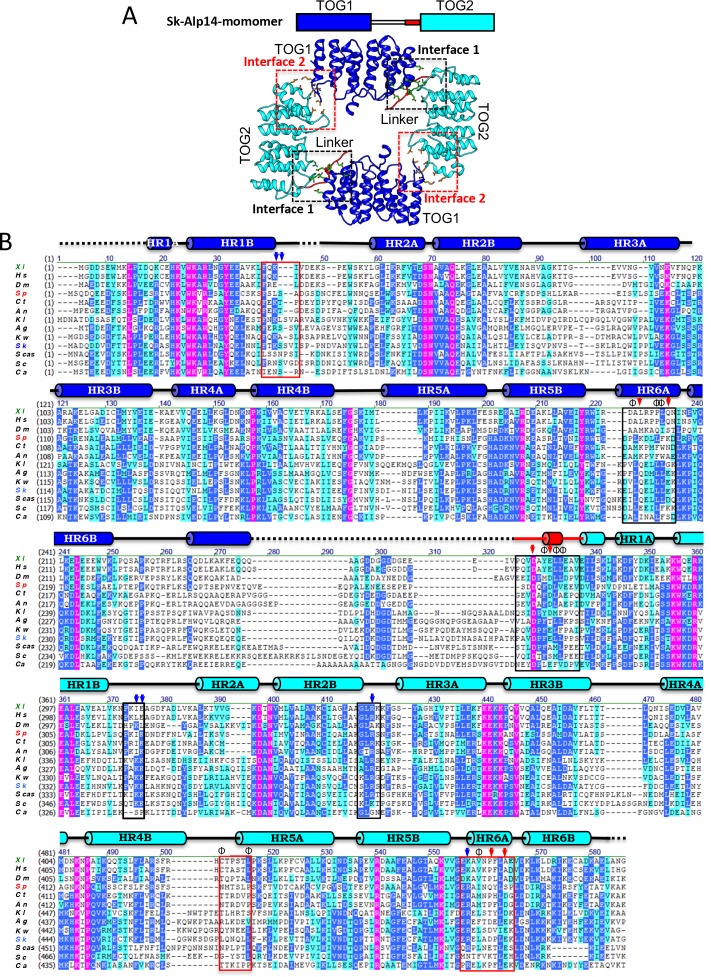
(A–B) 3.6 Å X-ray crystal structure of the S. kluyveri 1:2:2 sk-Alp14:αβ-tubulin:DRP reveals pseudo-dimeric head-to-tail subunits (red and orange) in a TOG square assembly consisting of four TOG domains bound to four αβ-tubulins (α-tubulin shown in cyan and β-tubulin shown in green) in a wheel-like organization. (A) Structure with DRP (yellow) bound to each αβ-tubulin. (B) Structure with DRP computationally removed. Each αβ-tubulin (α1β1) is positioned 90° rotated from its polymer-forming interface on its neighboring αβ-tubulin (α2β2). (C) Pseudo-dimeric TOG1-TOG2 subunits, shown in orange and red, respectively, form a head-to-tail TOG square (inset). Interface 1 is formed by the N-terminus of TOG2 and the TOG1-TOG2 linker binding to the C-terminus of the TOG1 domain of a second subunit, forming a 90° corner. Interface 2 is formed by the N-terminus of TOG1 binding the C-terminus of TOG2 within the same subunit in a 90° corner (Figure 2—figure supplement 1I). (D) Rainbow view of TOG1-TOG2 with N- and C-termini displayed in a blue-to-red color gradient, while the other subunit is displayed in grey. Each TOG is composed of six HEAT repeats (numbered). (E) Close-up view of interface 1. A hydrophobic zone stabilizes interface1 (yellow and highlighted by red outline) involving Leu220 (L220) and Leu217 (L217) of the TOG1 inter-HEAT 5–6 loop, Leu179 (L179) of the HEAT 6 A-helix in TOG1 (red ribbon) stabilized by linker residues (solid orange) Phe302 (F302), Leu304 (L304), and Leu305 (L305). An ionic zone guides interface 1 involving Glu219 (E219) of TOG1 inter-HEAT 5–6 loop and Glu305 (E305) of the TOG1-TOG2 linker, forming salt bridges with Lys346 (K346) and Lys347 (K347) of the TOG2 (light orange) inter-HEAT 1–2 loop and Arg390 (R390) of the TOG2 HEAT 2,3 loop, respectively. (F) Close-up view of interface 1, as in C, displaying residue conservation based on the alignment shown in Figure 2—figure supplement 2. (G) Close-up view of interface 2. A hydrophobic zone stabilizes interface 1 involving Leu477 (L477) and Leu472 (L472) of the TOG2 inter-HEAT4-5 loop with Ile43 of the TOG1 inter-HEAT1-2 loop. Ionic zone selectively guides interface 2, involving Lys39 (K39) and Ser41 (S41) of the TOG1 inter-HEAT1-2 loop and helix 1B with Arg517 (R517), Glu518 (E518), Asp521 (D521), and Glu524 (E524) of the TOG2 inter-HEAT5-6 loop and A-helix. (H) Close-up view of interface 2, as in D, displaying reside conservation based on the alignment in Figure 2—figure supplement 2.