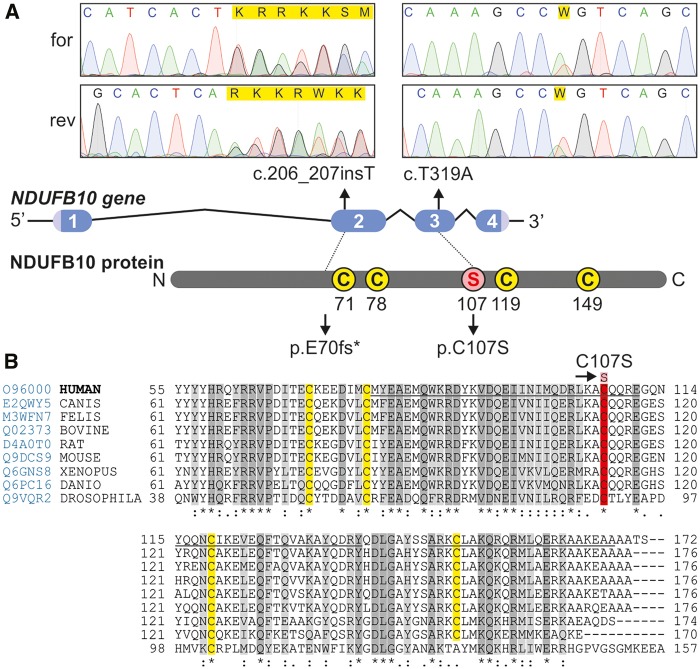
Figure 4.
Mutations in NDUFB10 and sequence alignment over multiple species. The position of the mutations in NDUFB10 is shown on a diagram of the gene, and respective Sanger sequencing results from the forward (for) and reverse (rev) sequencing are shown. The homology of NDUFB10 over multiple species (Homo sapiens, Canis lupus, Felis catus, Bos taurus, Rattus norvegicus, Mus musculus, Xenopus laevis, Danio rerio, Drosophila melanogaster) was built using Clustal Omega available online at uniprotkb, accessed at www.uniprot.org/align on 7/16/2016. Prediction of secondary structure helical motifs (underlined regions) was done according to Jpred4 prediction software accessed at www.compbio.dundee.ac.uk/jpred4 on 7/16/2016. Highly conserved amino acids are shown in dark grey and moderately conserved amino acids in light grey, whereas in yellow are shown the cysteines of the presumptive CXnC structures. The mutated amino acid C107S is shown in red.