Figure 1:
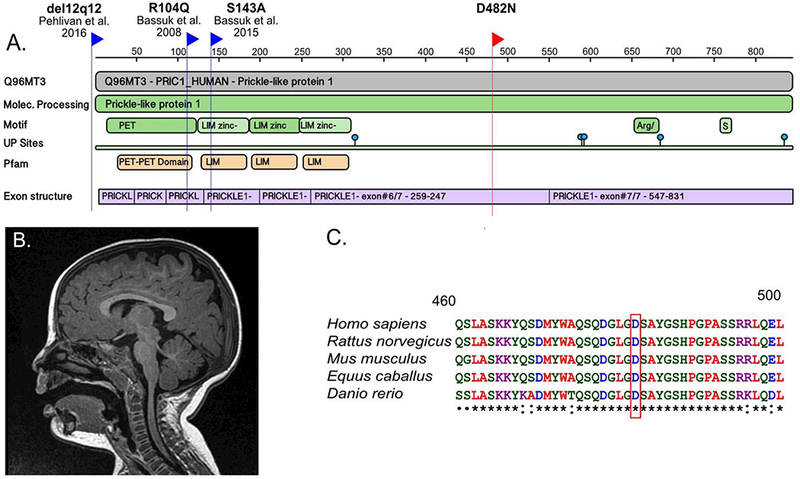
(a) Protein Feature View for PRICKLE1 from the RCSB Protein Data Bank (PDB) website. The blue flags indicate the location of human PRICKLE1 mutations that have been previously reported, including a de novo mutation associated with fetal agenesis of the corpus callosum and polymicrogyria (Bassuk & Sherr, 2015), a homozygous recessive mutation associated with myoclonic epilepsy (Bassuk et al., 2008), and a ~30-kb deletion involving the 5’ untranslated region and start of the PRICKLE1 coding sequence associated with Charcot Marie Tooth Disease and epilepsy (Pehlivan et al., 2015). The red flag indicates the novel de novo PRICKLE1 missense mutation. (b) Midline sagittal image from an MRI conducted at 1-year-old showing normal findings. (c) Multiple species protein alignment of the region surrounding the amino acid change caused by the novel de novo mutation in PRICKLE1. The red box indicates the reference amino acid, D, in humans and other vertebrates. The amino acid is completely conserved in vertebrates used in the alignment, and is within a highly conserved domain across all species. Peptide sequences were downloaded from UniProt BLAST and the alignment was performed using the Clustal Omega website (http://www.ebi.ac.uk/Tools/msa/clustalo/).
