Abstract
Canalization, or robustness to genetic or environmental perturbations, is fundamental to complex organisms. While there is strong evidence for canalization as an evolved property that varies among genotypes, the developmental and genetic mechanisms that produce this phenomenon are very poorly understood. For evolutionary biology, understanding how canalization arises is important because, by modulating the phenotypic variation that arises in response to genetic differences, canalization is a determinant of evolvability. For genetics of disease in humans and for economically important traits in agriculture, this question is important because canalization is a potentially significant cause of missing heritability that confounds genomic prediction of phenotypes. We review the major lines of thought on the developmental-genetic basis for canalization. These fall into two groups. One proposes specific evolved molecular mechanisms while the other deals with robustness or canalization as a more general feature of development. These explanations for canalization are not mutually exclusive and they overlap in several ways. General explanations for canalization are more likely to involve emergent features of development than specific molecular mechanisms. Disentangling these explanations is also complicated by differences in perspectives between genetics and developmental biology. Understanding canalization at a mechanistic level will depend on conceptual and methodological approaches that integrate quantitative genetics and developmental biology.
Keywords: Robustness, canalization, nonlinearity, epistasis, genotype-phenotype maps
1. Introduction
“The form of a living plant or animal is continuously kept in being in spite of the fact that material is passing all the time through the system.”
Waddington, 1957, pg. 2
In a series of experiments in the 1940s and 50s, Waddington demonstrated apparent acquired inheritance of the ether-induced bithorax phenotype in fruit flies [1–7]. He explained this finding as a consequence of selection acting on developmental processes that stabilize the phenotype in a particular environmental context. This tendency to buffer either genetic or environmental perturbations is known as canalization and is central to the evolution of complex life [1, 8–10]. Variation is necessary for evolution to occur. With increasing complexity, that variation has increasing potential for deleterious effects. As the number of interacting parts and processes increases, the ability to alter any one part without adverse effects on others will decrease. It is not surprising, therefore, that the genomes of healthy humans harbour large numbers of deleterious mutations of varying severity [11]. However, these variants do not result in disease because their effects are buffered. How this occurs is a long-standing question to which there is not yet a satisfactory answer. Recent work, however, is beginning to shed light on this question. This essay will discuss the current state of knowledge of the developmental basis for canalization and place this in the context of complex trait genetics and evolutionary developmental biology.
The concept of canalization is closely tied to Waddington’s metaphoric epigenetic landscape and his concept of epigenetics. He used all three of these terms in two very different contexts. These are the differentiation of cells and tissue types on the one hand and the suppression of phenotypic variation among individuals on the other. Waddington saw the stabilization of alternative cell fates, for example, and the modulation of phenotypic variance of a particular trait in a population, as different expressions of the same underlying phenomenon at different scales. In evolutionary biology, most interest has focused on the modulation of phenotypic variance at the population level [8]. In parallel, however, numerous studies in developmental biology invoke canalization in analyses of cell fate specification [12–15]. These applications of Waddington’s concepts are united by a view of development that is probabilistic or stochastic, invoking the need for mechanisms to impose order on an otherwise disordered process of development [16]. Beyond this broad similarity, though, it is not evident that the mechanisms that buffer phenotypic variation at the among-individual level and those that stabilize cell and tissue fates are in any way related.
Even so, the fact that Waddington saw the modulation of phenotypic variation and the stabilization of pathways of developmental differentiation as part and parcel of the same phenomenon is revealing. Waddington clearly thought of development as inherently stochastic. That is, through its multiple levels of organization and complex, interacting components, cells, tissues and organisms are capable of expressing tremendous variation, the overwhelming majority of which is deleterious [7]. In Waddington’s view, these influences flow through levels of organization from the biochemical basis of developmental processes to gross anatomical features [17]. This rich, multi-level, view of development presaged systems biology but also contrasts with current systems approaches, such as the gene regulatory network metaphor, that represent developmental systems at a particular phenomenological slice – that of genes, gene interactions and cis-regulatory sites [18]. Waddington argued that systems capable of so much variation must have evolved mechanisms to suppress it [7]. He also argued that such mechanisms might exist at several levels of development and that some might be embedded features of developmental systems rather than specific, dedicated mechanisms.
More than seventy years after Waddington articulated his ideas, we still do not have a generalizable understanding of the mechanistic basis for canalization. In this essay, we focus only on the modulation of variation among individuals. We argue that explanations of canalization fall into two groups. The first emphasizes specific molecular mechanisms such as heat shock proteins [19, 20]. The second emphasizes more deeply embedded or emergent features of development. This latter category includes gene interactions and network redundancies, heterozygosity, and nonlinearities in developmental processes. These explanations for canalization are not exclusive, but the latter category is likely to yield general explanations.
2. Definitions and Related Concepts
“Another essential point about histogenesis (and morphogenesis) is that the degree of canalization is under genetic control. That is to say, individuals of some genotypes show a more powerful tendency to regulate to the normal canalised paths of development than do others.”
Waddington, 1957. P. 20
Canalization and several closely related terms are used in different and overlapping ways. To avoid confusion, we define these terms below and relate them to one another. Canalization, or the tendency to buffer variation, is about the modulation of phenotypic variance due to factors other than the genetic or environmental variance, per se. Phenotypic robustness is a more general concept that includes developmental stability and canalization [10, 21]. Whereas canalization refers to the minimization of variation among individuals, developmental stability is the tendency to minimize variation among replicated structures within individuals [22]. Developmental stability is most often measured via random, normally distributed departures from symmetry where symmetrical development is expected, or fluctuating asymmetry [23]. Canalization and developmental stability are partially overlapping phenomena. This is evident in the case of threshold or discrete traits where the combination of trait frequency and underlying developmental stability determine the frequency of asymmetric expression [24]. Palmer [25] has also argued for a connection between symmetry-breaking and large-scale morphological transitions, or evolutionary novelties. It is not known, however, to what extent measures of canalization and developmental stability capture the same biological phenomena.
The association of canalization and developmental stability has been studied by examining correlations of among-individual and fluctuating asymmetry variance across both traits within populations and across populations. In the latter situation, this comparison is only informative if genetic variance and environmental conditions are controlled. The studies of this issue are inconclusive, with some pointing to independence of these two variance components [26–28] while others show varying degrees of association [29–32]. We have recently compared FA and among-individual variances across both traits and genotypes in mice and found no significant relationship [33]. Taken together, these results suggest that the mechanisms driving variation in measures of canalization and developmental stability are, at least, partially distinct.
Phenotypic plasticity and the related norm of reaction overlap conceptually with canalization. Phenotypic plasticity is simply the tendency for phenotypic variation to be influenced by environment [34]. A reaction norm is a predictable relationship between some environmental factor and a phenotypic trait. These concepts are often seen as the opposite of canalization [34–36], but this is an over-simplification. To begin with, these concepts have distinct histories and motivations. While canalization was motivated by developmental biology, phenotypic plasticity has a more naturalistic or population biology origin. Coined by Woltereck [37], the concept of a reaction norm was fleshed out by Schmalhausen [38] whose primary interest was in the central role of stabilizing selection in evolution. Independently of Waddington, Schmalhausen proposed a concept very similar to canalization which, in the English translation, he termed autonomization [38]. For him, reaction norms, shaped by stabilizing selection, are adaptive. Accordingly, autonomization is not just any minimization of any variation. Rather, it is specifically the minimization of non-adaptive variation around the reaction norm [38]. This contrasts significantly with current thinking on plasticity. In the current consensus, plasticity is not necessarily adaptive [39, 40]. As Pigliucci et al. point out, under this definition, Waddington’s example of the bithorax phenotype induced by ether is actually a case of non-adaptive phenotypic plasticity [41]. Plasticilty can also be separated into active and passive, where passive refers to the variation minimzed by environmental canalization [42, 43].
The conceptual overlap of phenotypic plasticity and robustness or canalization matters, because distinguishing observations, patterns and process is critical to elucidating underlying mechanisms. Canalization and phenotypic plasticity are merely abstractions from patterns in data. They are not processes or mechanisms. As such, their limited explanatory value rests solely on the extent to which they represent groupings of patterns expressed by a set of dynamics. The statement, for example, that an observed reduction in variance is explained by canalization simply means that the observation groups with a set of similar patterns that we don’t understand but have chosen to label as canalization. If that grouping is not sensible, it is unlikely to map on to mechanisms in a tractable way. The problem with viewing canalization as simply the inverse of plasticity is that plasticity is a much more general phenomenon. Accordingly, the term canalization is likely to cover such a heterogeneous set of patterns that attempts to explain all in mechanistic terms are futile.
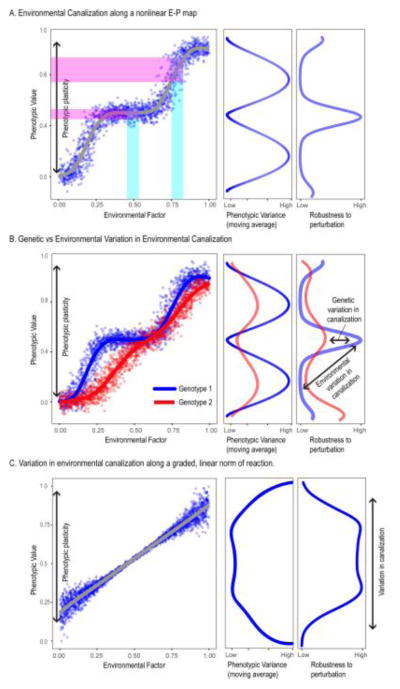
Wagner et al.’s [8] definition of canalization avoids this confusion. They define canalization as the suppression of phenotypic variation of either genetic or environmental origin. As they argue, this makes canalization a dispositional concept, referring to a tendency or potential. Accordingly, canalization is the tendency to suppress variation [8]. Canalization is not a component of an observed phenotypic variance. Rather, it is a component of variability, or the tendency to vary [8]. Observed differences in phenotypic variation are consequences of this tendency combined with individual circumstances such as the presence of perturbations or the amount of segregating genetic variation. Canalization is, therefore, only measured by observing differences in magnitudes of variation between samples or populations when controlling for factors such as the genetic variance and the magnitude of environmental effects as well as any known determinants of the trait in question such as age, sex or size [35]. Defined in this way, canalization is distinct from the more general concept of phenotypic plasticity (Figure 1). Importantly, this definition also allows for the possibility of changes in among-individual variance along the norm of reaction. Such changes in among-individual variance can result from a nonlinear norm of reaction curve [43] (Fig 1A). The shape of such norm of reaction curves can vary among genoytpes (Fig. 1B). However, they might also result from other factors such as destabilizing effects of environmental stress or genetic perturbations [44] (Fig 1C). Scharloo’s classic experiments with the Drosophila cubitus interruptus and hairless mutants showed that selection can alter the phenotypic responses to temperature variation [45, 46]. More recently, Debat et al.[47] showed that a series of 16 mutations influence sensitivity to temperature for Drosophila wing size and shape, while temperature also modulates the effects of those same mutations. These experiments nicely demonstrate variation in environmental canalization due to environment (along a norm of reaction) or genotype (modulating the norm of reaction) as well as the complex relationship between canalization and phenotypic plasticity.
Figure 1. Environmental canalization and phenotypic plasticity.
To illustrate the relationship between phenotypic plasticity and environmental canalization, we simulated a trait varying across a norm of reaction in response to an environmental effect. In each case, the phenotypic value is a function of the deterministic environmental effect plus an random term that is isotropic and normally distributed. In A, the variance of the random term is constant across the norm of reaction. However, a nonlinear relationship between the environmental effect and the phenotypic outcome generates variance heterogeneity across the norm of reaction. The downward modulation of variance across the middle of the range is the canalization effect. In B, two genotypes are shown that differ in the shape of the relationship between the environmental factor and the phenotypic outcome. This also generates a differential modulation of phenotypic variance. In this case, there is genetic variation in environmental canalization. In C, variance (the random term) increases away from the mean environmental value. This might occur, for example, if the more extreme environmental conditions are increasingly stressful, resulting in less stable development. In all cases, the entire range of phenotypic effects possible under different environmental conditions is the phenotypic plasticity. The shape of the curve that relates the phenotype to the environmental condition is the norm of reaction. The differential modulation of variance along and across these curves is environmental canalization. Environmental canalization can vary due to genotype or environment.
3. The Evidence for canalization
“It is a very general observation, to which little attention has been directed (but see Huxley, Plunkett and Ford), that the wildtype of an organism…, is much less variable in appearance than the majority of mutant races.”
Waddington, 1942, pg. 563
Although the origin of the canalization concept is linked to Waddington’s genetic assimilation experiments, the most general and convincing evidence for the phenomenon is the tendency for mutants to exhibit a larger range of variation than the wildtype. As the above quote from Waddington’s 1942 essay on genetic assimilation shows, the observation that mutants are more variable had been “in the ether” for some time before his seminal work. Schmalhausen also viewed the greater variability of mutants as accepted truism rather than an original observation [38]. In 50’s and 60s, geneticists documented increased among-individual variance within genotypes for a variety of mutations in Drosophila and mice [46, 48–51]. Figure 2 shows some examples of this phenomenon. Importantly, these early studies showed that the tendency for increased among-individual variance in mutants responded to selection, suggesting a heritable basis for canalization [49]. The responsiveness of phenotypic variance within genotype to selection has been confirmed more recently, perhaps most robustly in Arapidopsis [52]. In mice, our group has quantified significant heritability for phenotypic variance in 62 genotypes of isogenic mice derived from pairwise crosses among the eight collaborative cross founder strains [53].
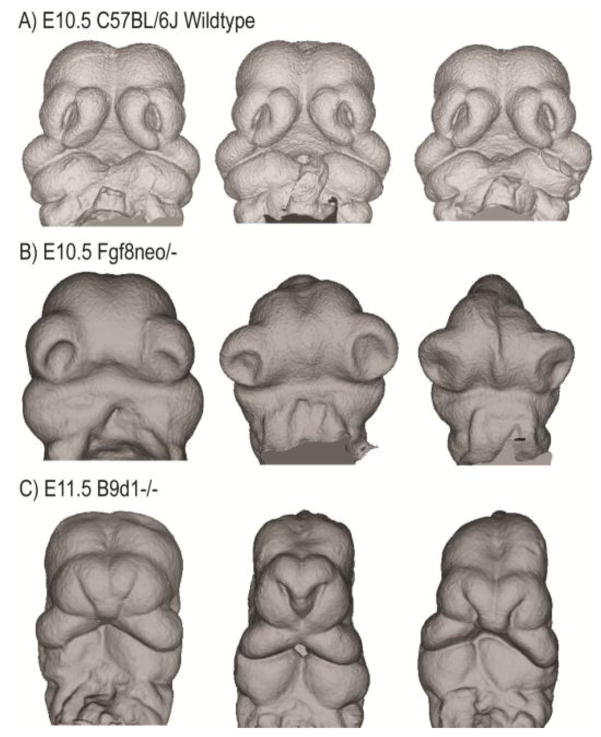
Figure 2. Variation among isogenic mutant mice with the same genotype.
MicroCT scans showing the front of the face of two different genotypes of embryos in comparison to controls. A) 10.5 day Wildtype C57BL6J mouse embryos. B) Fgf8neo/− (~80% loss) embryonic day 10.5 embryos (Green et al 2017). C) Examples of B9d1−/− (null) 11.5 day embryos.
In earlier work, our group compared phenotypic variances among mutants and wildtypes for craniofacial shape and found that mutations with large phenotypic effect tend strongly to be associated with increased phenotypic variance [54]. Similarly, a meta-analysis of studies in diverse mutations in nematodes revealed consistent increases in variance of comparable magnitude to mutational effects on the phenotypic mean [55]. In zebrafish, DeLaurier et al. found that a mutation in mef2ca dramatically elevates both the among-individual variance and fluctuating asymmetry [56]. While not all mutations affect both the variance and the mean (independently of statistical association of the mean and variance), it is clear that this frequently occurs, supporting the view that canalization effects are real phenomena in need of mechanistic explanation.
Another source of evidence for canalization is variable penetrance of mutations in natural populations. Variable expressivity for genetic disease in humans is well known and has received significant attention in recent years. Recent genomic work in humans has revealed the remarkable number of variants associated with major disease that are present in individuals who show no phenotypic effects of those diseases [57–59]. As part of their Resilience Project, Chen et al search for mutations in 874 genes believed to cause 584 distinct severe Mendelian childhood disorders in an aggregated cohort of over 500,000 genomes. In total, they identified 13 candidate resilient individuals spanning 8 diseases [57]. The analysis of aggregated reference human data sets such as the Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/) which excludes individuals affected by a severe pediatric disease provides a remarkable resource to explore genomic variation. Taraillo-Graovac et al interrogated the Exac database for rare pathogenic disease variants and found remarkably that 1,717 ExAC individuals (~2.8% of the ExAC population) harbored disease-associated variants. When combined with the recognized high degree of non-penetrance and highly variable expressivity of many rare diseases, these data do suggest the presence of biological mechanisms that mitigate against the impact of high penetrant mutations in humans.
4. Epistasis and Genetic Canalization
The effects of mutations often depend on genetic background. In a recent example using mice, Percival et al. showed that the craniofacial shape effects of three mutations in Sprouty genes, exhibit dramatically different shape effects when placed on three different inbred backgrounds [60]. In one case, the same mutation had diametrically opposite effects on two backgrounds. The Percival et al. study illustrates the broader significance of epistasis for complex traits and its relation to canalization. Interactions with other genes or background can alter the phenotypic effects of many, if not most, genetic variants. These epistatic effects may be to modulate penetrance, but they can also change the direction of the effect, the timing or locations of effects, or the range of pleiotropic effects [61, 62]. Epistasis is only relevant to canalization when it involves modulation of the magnitude of phenotypic effect. Although it is not known how frequently this occurs, it is likely to be quite common [62].
But what does it mean to posit epistasis as a cause of canalization? As Roff [34] has argued, the confounding of models and mechanisms has produced much confusion about canalization. The relationship between epistasis and canalization illustrates this perfectly. By Wagner’s [8] definition, genetic canalization is the suppression of the phenotypic effects of genetic variation. In his formalization, the genetic variance of a trait depends on both the frequency of the segregating alleles that influence it and the magnitudes of phenotypic effect of those alleles. Genetic canalization involves modulation of those phenotypic effects. All genetic canalization effects are attributable to epistasis in a quantitative genetic sense [63]. Yet large, population-level quantitative genetic effects are no assurance that underlying biological interactions between genes and/or gene products are understood or can be identified in individuals. To say epistasis explains genetic canalization in a mechanistic sense is therefore to commit Roff’s sin of confounding a quantitative model with a mechanistic explanation.
At the heart of this issue is the fact that quantitative genetics and developmental biology are concerned with different kinds of questions and have different definitions of ostensibly the same phenomena. While quantitative genetics is concerned with partitioning phenotypic outcomes into cumulative, statistically-defined phenomena, developmental biology is concerned with physical mechanisms such as specific molecular, cellular or tissue-level interactions. While the genetics of a population of clones is singularly uninteresting, its developmental biology may be fascinating. As a result, these fields produce different kinds of explanations. Quantitative genetics defines most phenomena at the population level and is fundamentally concerned with variation. The mechanisms of interest to developmental biology occur in individuals and, at least under the current paradigm, variation is more often a nuisance than a direct object of study. In genetics, epistasis is a statistical effect while in developmental biology it is generally viewed as a mechanistic interaction between gene products. The two versions of epistasis are linked conceptually but are not identical.
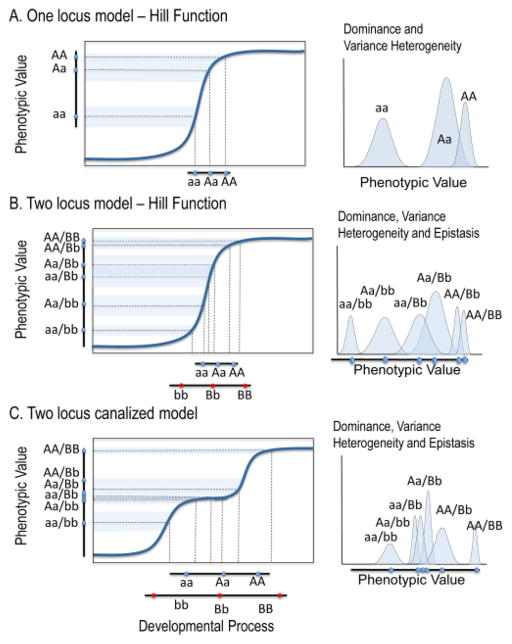
Canalization is a property of individuals that is almost always measurable only at a population level [8]. In genetic terms, a genetic factor increases canalization if, all else being equal, it reduces the phenotypic variance [35]. For genetic canalization, the influence of a gene on the phenotypic effect of other genes is, by definition a case of epistasis. Genetic variation in such effects could be described as differential epistasis. But does this mean that epistasis fully explains genetic canalization? Not necessarily. Gene interaction effects imply nonlinear effects on phenotype. However, this is a statistical model of variation and not a mechanistic description of actual physical interactions. Nonlinearities in development arise in myriad ways (see pg 15 below). If we start from the assumption that the relationship between some developmental factor and a phenotypic outcome is nonlinear, one can see that additive effects on the developmental factor result in both variance heterogeneity and gene interaction effects at the phenotypic level (Figure 3). This is demonstrated in the models of Sean Rice [63, 64] where variance heterogeneity emerges as a consequence of nonlinear genotype-phenotype maps. Similarly, genetic epistasis emerges as a consequence of nonlinearities in developmental processes that are influenced by multiple genes. Thus, depending on one’s perspective (genetic or developmental), epistasis can be seen as either a cause or a consequence of canalization.
Figure 3. Developmental nonlinearity and variance heterogeneity and epistasis.
This schematic shows how additive variation in a developmental process produces non-additive variation when that process has a nonlinear relationship to phenotypic outcomes. The first column shows genotype-phenotype maps while the second column shows phenotypic distributions by genotype. B and C show two of many possible two locus models. Both generate variance heterogeneity and epistasis.
5. Specific Mechanisms of Canalization
Mechanistic explanations for canalization have tended to fall into one of two categories. The first involves specific molecular mechanisms that have evolved to buffer variation. The second posits that canalization effects arise from embedded or emergent features of development. These explanations are not mutually exclusive, but they do imply very different views of biological systems.
5.1. Heat shock proteins and other molecular chaperones
Rutherford and Lindquist reported that perturbations to Hsp90, a heat shock protein resulted in an organism-wide increase in phenotypic variation [20]. Based on this and related work, Rutherford and others have argued for a general role for molecular chaperone proteins in buffering phenotypic variation [19, 65, 66]. The rationale for this idea is compelling. Chaperone proteins are involved in protein folding but are also implicated in stress responses and in re-folding damaged proteins [67]. It is plausible that perturbations to such proteins would impair stress response and increase the environmental variance as is demonstrated. The influence of variation in heat shock proteins on phenotypic variation has now been studied in a variety of systems. In Arabidopsis, Hsp90 is associated with variance heterogeneteity for loci influencing several quantitative traits [68, 69]. The same has also been shown for several different heat shock proteins in Drosophila [27], although other studies have shown weaker effects [70]. Interestingly, in yeast, mutations to Hsp90 may increase or decrease genetic canalization in a genotype-dependent manner [71]. In general, however, there is good evidence that perturbations to heat shock proteins associate with variance heterogeneity and modulation of environmental variance.
The problem with the idea that heat shock proteins are specific molecular mechanisms that act as master regulators of canalization or “capacitors of phenotypic variation,” is not a lack of evidence for their association with canalization. Rather, it is the abundant evidence that such associations occur for many other genes involving many different pathways. Analogous to chaperones, mutations in cohesins also appear to elevate variability. This makes sense because disruptions to chromatid structure can produce widespread disruption of gene expression. Random changes to gene expression and variable malformations in diverse organs, for example, occur in Nipbl mutants, a model for Cornelia de Lange syndrome [72–74]. A genome-wide increase in the variance of gene expression in mice also occurs as a result of a mutation to BAF155, a chromatin remodelin protein [75]. In mouse craniofacial development, varying degrees of effects on phenotypic variation are seen in a variety of genes including Papps2, Fgf8, and Ap-2 [76–78]. A similar increase in phenotypic variance occurs in histone mutations in yeast [79]. Finally, genome-wide studies of variance heterogeneity in Arabidopsis and Drosophila have identified multiple loci, but in genes unrelated to molecular chaperones such as a copper transporter factor [80] and genes in multiple other pathways [81]. These results might suggest that heat shock proteins have buffering roles that are specific to particular environmental conditions or organismal contexts and that there are other mechanisms that server other bufferering roles. Alternatively they may suggest that the increased variance that results from perturbations to these genes is an example of a more general phenomenon in development and that their association with canalization is reflective of their functional importance rather than their specific role in buffering variation.
5.2 Methylation
Another mechanism proposed to modulate phenotypic variance is methylation [82, 83]. The idea here is that regions of the genome are epigenetically differentiated in highly methylated or completely unmethylated states, stabilizing the expression of genes in those regions. A prediction from this hypothesis is that perturbations to genes that affect methylation patterns such as methyltransferases would impact canalization. Suggestive evidence for this idea comes from known cases of variation in penetrance to due variable methylation of IAP retrotransposon sites, or “meta-stable epialleles” [84]. While the classic example of this is the Agouti mouse [85], there are many other known cases. In A/WysnJ mice, for example, variable penetrance of cleft lip and reduced morphological integration is related to an IAP retrotransposon that influences Wnt9b expression [86–88].
A recent series of experiments by the Kimmel lab provides a fascinating example of this phenomenon in zebrafish. They observed that a mutation in the mef2ca gene produced a highly variable skeletal phenotype [56]. This gene is important in skeletal development and is involved in regulating the choice between ligament and bone in the zebrafish craniofacial complex. They determined that the penetrance of the mef2ca mutation was related to the proportion of cells directed to each of those two fates and that the degree of penetrance was heritable. In two lines that differed in penetrance, they determined that the heritable variation in penetrance was associated with to differential methylation of a nearby retrotransposon site that contains an enhancer for mef2ca [89]. This example is notable not just as an example of methylation as a mechanism for canalization but also because it combines the two senses in which Waddington used canalization and the epigenetic landscape – the reduction of phenotypic variance on the one hand and the specification of cell and tissue fates on the other.
5.3 MicroRNAs
MicroRNAs are small, non-coding RNAs that are involved in post-transcriptional regulation. They are numerous and diverse, constituting 1–2% of plant and animal genomes [90, 91]. MicroRNAs may contribute to canalization by buffering protein levels against variation in gene expression [92]. This may be particularly important for genes expressed in low and consequently relatively more variable levels, which is true for many genes with important development roles [93]. In a prominent example, Choi at al. [94] have revealed how miR-430 dampens and balances protein levels for the Tgf-β agonist squint and antagonist lefty in zebrafish. Also in zebrafish, loss of two miMRIs results in a two-fold increase in the phenotypic variance for histomorphometric traits in vascular endothelium [95]. Similarly, loss of miR-9 in Drosophila elevates phenotypic variation in the formation of sensory organs and in the expression of several key genes [96]. This study also reveals that the miRNA based regulation can produce a nonlinear relationship between mRNA and protein expression.
Clearly, it is plausible that microRNAs contribute to canalization. It is not known, however, to what extent they contribute to variation in canalization within population or at longer evolutionary time scales. Although recent work points to significant contributions by microRNAs to variation in complex traits [97], such studies have yet to test for associations with variance heterogeneity.
6. Emergent or Embedded Mechanisms
“It is important to realize that the comparatively simple orderliness of the epigenetic landscape – its restricted number of values with their branching-points and characteristic contours-is a property of a higher order dependent on an underlying network of interactions which is vastly more complicated.”
Waddington, 1957, pg. 34–35
The alternative to specific evolved mechanisms that modulate canalization is that the modulation of phenotypic variance occurs via emergent or embedded features of development rather than specific, dedicated mechanisms. In this case, canalization of a phenotypic trait is produced by the same genes and processes that underlie variation of that trait. As suggested by the above quote, this appears to have been Waddington’s intuition about the mechanisms of canalization. There are three lines of thought that fall into this category. One deals with properties of networks or systems. Another deals with excess production of proteins or other key resources in development and the third with the implications of nonlinearity in development.
6.1 Network Properties
Waddington described development as consisting of complex systems in which processes and interactions gravitated towards stable states [7]. In this, he appears to have inspired Rene Thom and the development of catastrophe theory [98], which has been highly influential in systems biology. Early attempts to model biological systems include the work of Stuart Kaufman and Stuart Newman on Boolean networks [99, 100] that contributed to the theoretical basis for gene regulatory networks [101]. How properties of systems resulted in canalization was a prominent theme in this early work in systems biology.
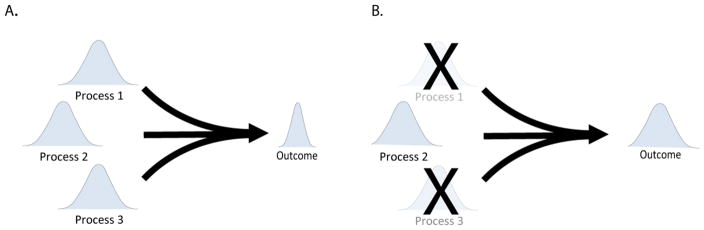
The most straightforward way in which embedded features of developmental systems might modulate variance is redundancy. As Soule [102] pointed out, variance of a process is a function of the number of independent inputs into that process and the variances of those inputs (Figure 4). In a process, such as growth, which depends on many (partially covarying) component processes, altering the variance of the component processes will change the variance of the ultimate outcome. For genes with redundant functions, null mutations might, therefore, be expected to produce a viable but more variable phenotype. Conversely, redundancy in gene function contributes to robustness to mutation in those genes [103, 104]. Exemplifying this effect, Morishita and Iwasa [105] show how multiple morphogen gradients can serve to stabilize positional information in Drosophila embryos. Here, multiple gradients effectively average out the noise variances of each gradient. In yeast, Levy and Siegal [106] showed that mutations to genes that are deeply embedded in interaction networks but are not redundant with other genes tends to disrupt phenotypic robustness (or variance heterogeneity). Conversely, mutations to hub genes that have functional redundancy with other genes tend not to affect robustness. This supports a role for functional redundancy in buffering.
Figure 4. Redundancy or Multiple Inputs and Canalization.
In A, variation in a hypothetical trait is determined by variation in three processes that vary independently of each other. The variance of the trait is lower than the variance of the component processes because of the averaging effect of multiple inputs. If any of the component processes are eliminated, such as might occur in a loss of function mutation for a redundant gene, the variance of the trait is increased.
Antagonistic gene interactions or regulatory feedback loops are the other major way in which network properties may contribute to canalization [99, 100]. Many examples of such interactions or feedback loops are known. As argued by Geiler-Samerotte et al. [107] in this volume, such interaction effects may have complex effects on the modulation of variance with the same mutation dampening or increasing phenotypic variance depending on the genetic context. In zebrafish, for example, the Schilling lab has shown how a feedback loop involving five genes stabilizes the retinoic acid morphogen gradient involved in establishing the antero-posterior body axis [108]. An obvious prediction from the regulatory feedback hypothesis is that perturbations to regulatory interactions would disrupt canalization. This prediction is nicely tested by Paulsen et al [109] in Xenopus. They show that negative feedback between BAMBI and SMAD7 stabilize BMP4 expression and that this, in turn, modulates phenotypic variance. A second prediction of this hypothesis is that epistasis contributes to variance heterogeneity. This is less well known, but epistasis has been associated with variance heterogeneity in yeast [110].
There is clearly good evidence that at least some network properties predicted to influence canalization do so in developmental systems. The current work in this area has only scratched the surface, however. In particular, it has proven immensely difficult to incorporate the role of interactions above the gene level, to paraphrase Waddington. Recent work advancing the ability to measure and manipulate physical forces in development is one key to this, but much needs to be done in order to incorporate cell and tissue-level phenomena into the molecular interactomes that are currently the raw material of systems biology [111]. Much work, therefore, needs to be done for the systems biology of robustness to be amenable to mechanistic explanation.
6.2 Heterozygosity
Lerner [112] argued that heterozygosity was the mechanistic basis for canalization. His argument rested on the premises that fitness peaks tend to be close to population means, that heterozygosity is greater at the mean than at phenotypic extremes, that inbreeding reduces fitness and increases variability, and that extreme phenotypes tend to be homozygotes for contributing genes. Much of this has subsequently been shown to be wrong. Empirical tests of Lerner’s hypothesis about a relationship between robustness and heterozygosity have shown weak and contradictory effects [113–118]. The stronger effects in these studies, in fact, point in the opposite direction and relate to changes in developmental stability and phenotypic variance in hybrid zones [119–122]. Most importantly, for polygenic traits, heterozygosity may not correlate with distance from the mean. Further, the reduction in fitness towards phenotypic extremes is generally due to recessive homozygotes and not to reduced heterozygosity [123]. However, Lerner’s thesis is not entirely without merit. In humans, Inbreeding increases the prevalence and penetrance of recessive disease, is associated with reduced birth weight and increased prevalence of birth defects [124]. It is also associated with increased prevalence of complex genetic disease [125]. The correlation between of consanguinity or inbreeding and variance heterogeneity, however, is not known and has not, to our knowledge, been addressed. Such a relationship would suggest a link between individual-level homozygosity and canalization. However, this correlation would likely be driven by the increased expression of rare, recessive variants than by some systemic effect of heterozygosity, per se.
6.3 Nonlinearities in Development
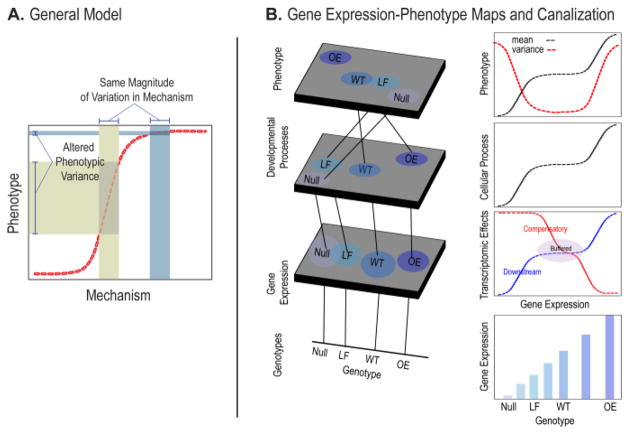
Nonlinearity is ubiquitous in biology. Often this is ignored for methodological convenience or theoretical necessity and not because the deviations from linearity are not of consequence [126]. Variance heterogeneity as a general consequence of nonlinear genotype-phenotype maps was shown by Sean Rice as part of his theoretical framework for integrating development and quantitative genetics [63, 127]. More recently, Morissey [128] has extended this theoretical framework for relating variation in development to phenotypic variation when the underlying relationships are highly nonlinear. This theoretical work shows that one consequence of nonlinearity in the mapping of phenotypic variation to underlying determinants such as gene expression or cellular processes is the modulation of phenotypic variation (Figure 5). This phenomenon has been suggested to be an explanation for variation in robustness of developmental systems [54, 63, 78, 129]. Ramler et al. have also proposed this as an explanation for modulation of robustness across a range of environmental conditions [130].
Figure 5. Developmental nonlinearity and phenotypic robustness.
A) A nonlinear curve relating variation in a mechanism (e.g. gene expression) to phenotypic outcome. The amount of phenotypic variation that corresponds to a constant amount of variation in the mechanism is dependent on the slope at each point along the curve. B shows an expansion of Lewontin’s Genotype-phenotype map in the first column and hypothetical gene expression to process and gene-expression to phenotype curves in the second. The number of levels is arbitrary in this example.
A closely related idea is the hypothesis that robustness derives from excess production of proteins [131]. The idea here is that above some threshold, the amount of a critical protein is sufficient and variation in protein level above this threshold will not affect the phenotype. This is the same argument made by Sewall Wright’s when addressing developmental basis of dominance [132]. This idea that excess developmental resources buffer against variation is also the basis for the much studied but weakly supported contention that phenotypic robustness decreases with nutritional stress [133, 134].
How does nonlinear mapping of gene expression to phenotype arise in development? This occurs in many ways and at different levels of development. In 1910, Hill [135] modeled the dissociation of oxygen and haemoglobin as a sigmoidal function The resulting Hill equation is widely used to characterize receptor-ligand relationships, which are generally nonlinear [136]. Transcriptional regulation involve feedback loops that can result in nonlinearity, and this has been proposed to contribute to genetic canalization [137]. Translational regulation may also be nonlinear as shown, for example, by the ribosome flow model [138]. Diffusion gradients for a morphogen can be nonlinear and the effect of morphogens within such gradients can have nonlinear effects because of their spatiotemporal patterning [139]. Nonlinearities can also arise at higher levels of mechanism. Zhang et al.’s multiscale model of limb development [140], for example, generates nonlinear process to phenotype mapping due to the nonlinear dynamics of cellular and tissue level processes.
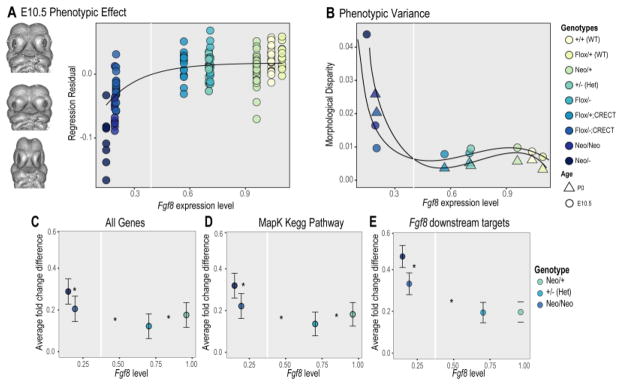
These nonlinearities occur at different scales and by different types of developmental mechanism (Figure 5). For nonlinearities to contribute to phenotypic robustness, their effects must be preserved across levels of mechanism to influence the mapping of genetic variation, gene expression or developmental processes to phenotypic variation. Our group has recently shown that this can occur [76]. Using a series of nine genotypes that vary in level of expression for Fgf8, a key gene for craniofacial development, we showed that the mapping of gene expression to phenotype is highly nonlinear. Loss of over 50% of the wildtype expression level is associated with minimal changes in phenotype while beyond this point, the phenotypic effects are large and increasingly severe. We modeled the predicted effects of this nonlinear gene-expression to phenotype map and found that the changes in phenotypic variance across the allelic series match our predictions (Figure 6). This shows that developmental nonlinearities can be preserved across the multiple levels of development to modulate phenotypic robustness.
Figure 6. The Fgf8 gene-expression phenotype map and robustness.
A) Gene-expression to phenotype map for an allelic series of E10.5 mouse embryos that vary in level of Fgf8 expression. The hypothetical genotypes are null, loss of function (LF), wildtype (WT) and overexpression (OE). This example shows a buffered range around the wildtype although many shapes of such curves are possible.
In the Fgf8 experiment, we extended variation in Fgf8 expression well beyond the range expected in natural populations. This begs the question of how such nonlinearities evolve. The local flattening of the gene-expression to phenotype around the wildtype could evolve by stabilizing selection acting on gene-interaction effects [8]. For a single trait, such effects are the same as variance heterogeneity or variance QTLs. If directional selection moves the phenotype along the curve, the slope of the genotype-phenotype realized within the population will change. This would alter heritability and, in turn, influence selection across generations. The question of how stabilizing or directional selection influence variational properties such as canalization has been an area of significant debate [141, 142]. Combining quantitative genetic models and theory with experimental manipulation of actual genotype-phenotype maps may provide answers to this important question.
7. Future Directions: Integrating Developmental Biology and Genetics
Recent years have seen a resurgence of interest in canalization and robustness. This resurgence appears to be driven from several independent trends. One is the discovery of new molecular mechanisms such as microRNAs and the resulting thought about their potential roles [143]. Another is the search for explanations for the missing heritability in complex traits [62]. A third is ongoing progress in systems biology that is leading investigators to tackle increasingly complex, emergent properties in quantitative models [144]. The resultant diversity of approaches and perspectives has advanced understanding of robustness in biology. However, diversity, often originating in different disciplines, has also created a situation reminiscent of the parable of the blind men and the elephant. Each tends to be concerned with a particular aspect of the problem, making it difficult to contextualize this progress in terms of a more general understanding of the mechanisms of canalization in development.
One consequence of this problem is that the mechanistic explanations that have been proposed for canalization are neither mutually exclusive nor conceptually distinct. The fact that disruptions to heat shock proteins may increase variation is not inconsistent with network properties or nonlinearities as explanations for canalization. In fact, both may apply to the same trait. Further, some explanations deal with overlapping phenomena. Antagonistic feedback loops and redundancies imply gene interaction effects and both are associated with nonlinearities. MicroRNAs may influence robustness via feedback, nonlinearity and redundancy. Heat shock proteins may influence robustness in a genetic-background dependent manner, linking the chaperone hypothesis and epistasis [71]. We have argued that some of this confusion can be traced to the difference in perspective between genetics and developmental biology and the resulting difficulty in translating concepts like epistasis from one to the other. Just as important, though, is the sheer complexity of biological systems and the myriad ways such systems might modulate the translation of genetic to phenotypic variation.
If the mechanistic basis for canalization is tractable, successful explanation must involve bridging across perspectives, mechanistic levels and expertise domains. Imagine the task of explaining robustness in a simpler, designed system like a car instead of an evolved complex organism. Some features, like shock absorbers might function to dampen specific environmental influences (bumps on the road). Other features, such as the design of the engine, ability to accelerate or turn or the skill of the driver might also contribute to robustness to perturbations like unexpected traffic, weather conditions or the mechanical failure of some component. These latter features are not separate from the integral design of the car but no less relevant. Currently, systems approaches to canalization tend to represent a particular phenomenological level, such as gene-regulatory networks. Although the need to break out of this mold is recognized by leaders in that field [18, 111], and there are ongoing efforts to place gene-regulatory networks into spatiotemporal contexts [145], this is very difficult to achieve.
Another promising approach is to combine imaging, morphometrics and developmental biology to study the quantitative origin of variation in development. The Fgf8 and Shh studies from our group are early examples of this approach [76, 146]. Here, the idea is to use the genotype-phenotype map paradigm [147] to construct gene-expression to phenotype maps in which multiple imaging, molecular and bioinformatic tools are used to quantitatively relate variation across multiple levels of mechanism. This approach also connects genetics and developmental biology in that it places a central focus on how variation is transmitted across the developmental mechanisms that translate genotype to phenotype. Recent advances in 3D imaging, particularly at the scale of whole embryos, and the ability to measure phenomena such as mechanical forces at the cellular and tissue level make this kind of approach increasingly promising [148–153].
Canalization is a fundamental property of complex organisms relevant to both evolutionary biology and the genetics of complex traits. We have reviewed past and current thinking on the mechanistic basis for this phenomenon. Canalization is unlikely to be due to a single mechanism, even in a particular biological context. Rather, the modulation of variance likely reflects the effects of multiple mechanisms, some of which might be specifically evolved to restrict the range of phenotypic outcomes, while others are embedded or emergent features of development. Understanding their relative contributions will depend on advances in systems biology that allow for multi-scale integration of imaging and molecular data as well as the development of theoretical models to make sense of the large and complex datasets that such approaches generate. As such approaches develop and mature in coming years, they are likely to enrich our understanding of the mechanistic basis for robustness. In so doing, this work will significantly illuminate the developmental-genetics of complex traits.
Acknowledgments
Funding: This work was supported by grants NIH R01 2R01DE019638 and NSERC 238992-17.
Footnotes
Conflict of Interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Waddington CH. The canalisation of development and the inheritance of acquired characters. Nature. 1942;150:563. [Google Scholar]
- 2.Waddington CH. Selection of the genetic basis for an acquired character. Nature. 1952;169(4302):625–6. doi: 10.1038/169625b0. [DOI] [PubMed] [Google Scholar]
- 3.Waddington CH. The genetic assimilation of an acquired character. Evolution. 1953;7:118–126. [Google Scholar]
- 4.Waddington CH. On a case of quantitative inheritence on either side of the wild-type. Z in Abstammungs u Vererb Lehre. 1955;87:208–28. doi: 10.1007/BF00309517. [DOI] [PubMed] [Google Scholar]
- 5.Waddington CH. Genetic assimilation of the bithorax phenotype. Evolution. 1956;10:1–13. [Google Scholar]
- 6.Waddington CH. The genetic basis of the ‘assimilated bithorax’ stock. J Genet. 1956;55:241–245. doi: 10.1007/BF02729015. [DOI] [PubMed] [Google Scholar]
- 7.Waddington CH. The Strategy of the Genes. MacMillan Company; New York: 1957. [Google Scholar]
- 8.Wagner GP, Booth G, Bagheri-Chaichian H. A population genetic theory of canalization. Evolution. 1997;51(2):329–347. doi: 10.1111/j.1558-5646.1997.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 9.Scharloo W. Canalization - Genetic and Developmental Aspects. Annual Review of Ecology and Systematics. 1991;22:65–93. [Google Scholar]
- 10.de Visser JA, Hermisson J, Wagner GP, Ancel Meyers L, Bagheri-Chaichian H, Blanchard JL, Chao L, Cheverud JM, Elena SF, Fontana W, Gibson G, Hansen TF, Krakauer D, Lewontin RC, Ofria C, Rice SH, von Dassow G, Wagner A, Whitlock MC. Perspective: Evolution and detection of genetic robustness. Evolution. 2003;57(9):1959–72. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 11.Henn BM, Botigue LR, Bustamante CD, Clark AG, Gravel S. Estimating the mutation load in human genomes. Nat Rev Genet. 2015;16(6):333–43. doi: 10.1038/nrg3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Méndez A, Mendoza L. A Network Model to Describe the Terminal Differentiation of B Cells. PLOS Computational Biology. 2016;12(1):e1004696. doi: 10.1371/journal.pcbi.1004696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moris N, Pina C, Arias AM. Transition states and cell fate decisions in epigenetic landscapes. Nat Rev Genet. 2016;17(11):693–703. doi: 10.1038/nrg.2016.98. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Zhang K, Xu L, Wang E. Quantifying the Waddington landscape and biological paths for development and differentiation. Proc Natl Acad Sci U S A. 2011;108(20):8257–62. doi: 10.1073/pnas.1017017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J, Lin F, Zhang X, Tanavde V, Zheng J. NetLand: quantitative modeling and visualization of Waddington’s epigenetic landscape using probabilistic potential. Bioinformatics. 2017;33(10):1583–1585. doi: 10.1093/bioinformatics/btx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert SF. Epigenetic landscaping: Waddington’s use of cell fate bifurcation diagrams. Biology and Philosophy. 1991;6(2):135–154. [Google Scholar]
- 17.Waddington CH. Towards a theoretical biology. Nature. 1968;218(5141):525–7. doi: 10.1038/218525a0. [DOI] [PubMed] [Google Scholar]
- 18.Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proceedings of the National Academy of Sciences. 2008;105(51):20063–20066. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutherford SL. From genotype to phenotype: buffering mechanisms and the storage of genetic information. Bioessays. 2000;22(12):1095–105. doi: 10.1002/1521-1878(200012)22:12<1095::AID-BIES7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 20.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396(6709):336–42. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 21.Wagner A. Robustness and evolvability in living systems. Princeton university press; 2013. [Google Scholar]
- 22.Hallgrimsson B, Willmore K, Hall B. Canalization, developmental stability, and morphological integration in primate limbs. Yearbook of Physical Anthropology. 2002;45:131–158. doi: 10.1002/ajpa.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Valen L. A Study of Fluctuating Asymmetry. Evolution. 1962;16:125–142. [Google Scholar]
- 24.Hallgrimsson B, Donnabhain BO, Blom DE, Lozada MC, Willmore KT. Why are rare traits unilaterally expressed?: trait frequency and unilateral expression for cranial nonmetric traits in humans. American Journal of Physical Anthropology. 2005;128(1):14–25. doi: 10.1002/ajpa.20187. [DOI] [PubMed] [Google Scholar]
- 25.Palmer AR. Asymmetry Breaking and the Evolution of Development. Science. 2004;306(828):833. doi: 10.1126/science.1103707. [DOI] [PubMed] [Google Scholar]
- 26.Debat V, Alibert P, David P, Paradis E, Auffray JC. Independence between developmental stability and canalisation in the skull of the house mouse. Proc Roy Soc Lond B. 2000;267:423–430. doi: 10.1098/rspb.2000.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi KH, Rako L, Takano-Shimizu T, Hoffmann AA, Lee SF. Effects of small Hsp genes on developmental stability and microenvironmental canalization. BMC Evol Biol. 2010;10:284. doi: 10.1186/1471-2148-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breno M, Leirs H, Van Dongen S. No relationship between canalization and developmental stability of the skull in a natural population of Mastomys natalensis (Rodentia: Muridae) Biological Journal of the Linnean Society. 2011;104(1):207–216. [Google Scholar]
- 29.Willmore K, Klingenberg C, Hallgrimsson H. Congruence between canalization and developmental stability in Macaca mulatta crania. American Journal of Physical Anthropology. 2005:224. [Google Scholar]
- 30.Santos M, Iriarte PF, Cespedes W. Genetics and geometry of canalization and developmental stability in Drosophila subobscura. BMC Evol Biol. 2005;5:7. doi: 10.1186/1471-2148-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breuker CJ, Patterson JS, Klingenberg CP. A single basis for developmental buffering of Drosophila wing shape. PLoS ONE. 2006;1:e7. doi: 10.1371/journal.pone.0000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padro J, Carreira V, Corio C, Hasson E, Soto IM. Host alkaloids differentially affect developmental stability and wing vein canalization in cactophilic Drosophila buzzatii. J Evol Biol. 2014;27(12):2781–97. doi: 10.1111/jeb.12537. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez PN, Lotto FP, Hallgrimsson B. Canalization and developmental instability of the fetal skull in a mouse model of maternal nutritional stress. Am J Phys Anthropol. 2014;154(4):544–53. doi: 10.1002/ajpa.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roff DA. Evolutionary Quantitative Genetics. Chapman & Hall; New York: 1997. [Google Scholar]
- 35.Flatt T. The evolutionary genetics of canalization. Quarterly Review of Biology. 2005;80(3):287–316. doi: 10.1086/432265. [DOI] [PubMed] [Google Scholar]
- 36.Stearns SC, Kawecki TJ. Fitness sensitivity and the canalization of life-history traits. Evolution. 1994;48:1438–1450. doi: 10.1111/j.1558-5646.1994.tb02186.x. [DOI] [PubMed] [Google Scholar]
- 37.Woltereck R. Weitere experimenelle Untersuchungen uber Artveranderung, speziell uber des Wesen quantitativer Artunterschiede bei Daphniden. Ver Deutsche Zool Gesell. 1909;19:110–172. [Google Scholar]
- 38.Schmalhausen II. Factors of Evolution. University of Chicago Press; Chicago: 1949. [Google Scholar]
- 39.Scheiner SM, Holt RD. The genetics of phenotypic plasticity. X. Variation versus uncertainty. Ecology and Evolution. 2012;2(4):751–767. doi: 10.1002/ece3.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stearns SC. The evolutionary significance of phenotypic plasticity. BioScience. 1989;39(7):436–445. [Google Scholar]
- 41.Pigliucci M, Murren CJ, Schlichting CD. Phenotypic plasticity and evolution by genetic assimilation. Journal of Experimental Biology. 2006;209(12):2362–2367. doi: 10.1242/jeb.02070. [DOI] [PubMed] [Google Scholar]
- 42.Forsman A. Rethinking phenotypic plasticity and its consequences for individuals, populations and species. Heredity. 2014;115:276. doi: 10.1038/hdy.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liefting M, Hoffmann AA, Ellers J. PLASTICITY VERSUS ENVIRONMENTAL CANALIZATION: POPULATION DIFFERENCES IN THERMAL RESPONSES ALONG A LATITUDINAL GRADIENT IN DROSOPHILA SERRATA. Evolution. 2009;63(8):1954–1963. doi: 10.1111/j.1558-5646.2009.00683.x. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi KH, Daborn PJ, Hoffmann AA, Takano-Shimizu T. Environmental Stress-Dependent Effects of Deletions Encompassing Hsp70Ba on Canalization and Quantitative Trait Asymmetry in Drosophila melanogaster. PLOS ONE. 2011;6(4):e17295. doi: 10.1371/journal.pone.0017295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scharloo W. The influence of selection and temperature on a mutant character (ciD) in Drosophila melanogaster. Arch Neerl Zool. 1962;14:431–512. [Google Scholar]
- 46.Scharloo W. Mutant expression and canalization. Nature. 1964;203:1095–1096. doi: 10.1038/2031095b0. [DOI] [PubMed] [Google Scholar]
- 47.Debat V, Debelle A, Dworkin I. Plasticity, canalization, and developmental stability of the Drosophila wing: joint effects of mutations and developmental temperature. Evolution. 2009;63(11):2864–76. doi: 10.1111/j.1558-5646.2009.00774.x. [DOI] [PubMed] [Google Scholar]
- 48.Rendel JM. CANALIZATION OF THE SCUTE PHENOTYPE OF DROSOPHILA. Evolution. 1959;13(4):425–439. [Google Scholar]
- 49.Rendel JM. Canalization and gene control. Logos Press; London: 1967. [Google Scholar]
- 50.Dun RB, Fraser AS. Selection for an Invariant Character - Vibrissa Number - in the House Mouse. Nature. 1958;181(4614):1018–1019. doi: 10.1038/1811018a0. [DOI] [PubMed] [Google Scholar]
- 51.Mather K. Genetical control of stability in development. Heredity. 1953;7:297–336. [Google Scholar]
- 52.Hall MC, Dworkin I, Ungerer MC, Purugganan M. Genetics of microenvironmental canalization in <em>Arabidopsis thaliana</em>. Proceedings of the National Academy of Sciences. 2007;104(34):13717–13722. doi: 10.1073/pnas.0701936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez PN, Pavlicev M, Mitteroecker P, Pardo-Manuel de Villena F, Spritz RA, Marcucio RS, Hallgrimsson B. Genetic structure of phenotypic robustness in the collaborative cross mouse diallel panel. J Evol Biol. 2016;29(9):1737–51. doi: 10.1111/jeb.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hallgrimsson B, Jamniczky H, Young NM, Rolian C, Parsons TE, Boughner JC, Marcucio RS. Deciphering the Palimpsest: Studying the Relationship Between Morphological Integration and Phenotypic Covariation. Evol Biol. 2009;36(4):355–376. doi: 10.1007/s11692-009-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baer CF. Quantifying the decanalizing effects of spontaneous mutations in rhabditid nematodes. Am Nat. 2008;172(2):272–81. doi: 10.1086/589455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeLaurier A, Huycke TR, Nichols JT, Swartz ME, Larsen A, Walker C, Dowd J, Pan L, Moens CB, Kimmel CB. Role of mef2ca in developmental buffering of the zebrafish larval hyoid dermal skeleton. Developmental Biology. 2014;385(2):189–199. doi: 10.1016/j.ydbio.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, Zhou H, Tian L, Prakash O, Lemire M, Sleiman P, Cheng WY, Chen W, Shah H, Shen Y, Fromer M, Omberg L, Deardorff MA, Zackai E, Bobe JR, Levin E, Hudson TJ, Groop L, Wang J, Hakonarson H, Wojcicki A, Diaz GA, Edelmann L, Schadt EE, Friend SH. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34(5):531–8. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- 58.Tarailo-Graovac M, Zhu JYA, Matthews A, van Karnebeek CDM, Wasserman WW. Assessment of the ExAC data set for the presence of individuals with pathogenic genotypes implicated in severe Mendelian pediatric disorders. Genet Med. 2017;19(12):1300–1308. doi: 10.1038/gim.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome C. Aggregation, Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Percival CJ, Marangoni P, Tapaltsyan V, Klein O, Hallgrímsson B. The Interaction of Genetic Background and Mutational Effects in Regulation of Mouse Craniofacial Shape. G3: Genes|Genomes|Genetics. 2017;7(5):1439–1450. doi: 10.1534/g3.117.040659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nadeau JH. Modifier genes in mice and humans. Nature Reviews Genetics. 2001;2(3):165. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- 62.Mackay TFC. Epistasis and Quantitative Traits: Using Model Organisms to Study Gene-Gene Interactions. Nature reviews Genetics. 2014;15(1):22–33. doi: 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rice S. The evolution of canalization and the breaking of von Baer’s laws: Modeling the evolution of development with epistasis. Evolution. 1998;52(3):647–656. doi: 10.1111/j.1558-5646.1998.tb03690.x. [DOI] [PubMed] [Google Scholar]
- 64.Rice SH. Theoretical approaches to the evolution of development and genetic architecture. Annals of the New York Academy of Sciences. 2008;1133:67–86. doi: 10.1196/annals.1438.002. [DOI] [PubMed] [Google Scholar]
- 65.Rutherford SL. Between genotype and phenotype: protein chaperones and evolvability. Nature Reviews Genetics. 2003;4:263. doi: 10.1038/nrg1041. [DOI] [PubMed] [Google Scholar]
- 66.Zabinsky RA, Mason GA, Queitsch C, Jarosz DF. It’s not magic - Hsp90 and its effects on genetic and epigenetic variation. Seminars in Cell and Developmental Biology. 2018 doi: 10.1016/j.semcdb.2018.05.015. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marques C, Guo W, Pereira P, Taylor A, Patterson C, Evans PC, Shang F. The triage of damaged proteins: degradation by the ubiquitin-proteasome pathway or repair by molecular chaperones. The FASEB Journal. 2006;20(6):741–743. doi: 10.1096/fj.05-5080fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sangster TA, Salathia N, Undurraga S, Milo R, Schellenberg K, Lindquist S, Queitsch C. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc Natl Acad Sci U S A. 2008;105(8):2963–8. doi: 10.1073/pnas.0712200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Samakovli D, Thanou A, Valmas C, Hatzopoulos P. Hsp90 canalizes developmental perturbation. J Exp Bot. 2007;58(13):3513–24. doi: 10.1093/jxb/erm191. [DOI] [PubMed] [Google Scholar]
- 70.Debat V, Milton CC, Rutherford S, Klingenberg CP, Hoffmann AA. Hsp90 and the quantitative variation of wing shape in Drosophila melanogaster. Evolution. 2006;60(12):2529–38. [PubMed] [Google Scholar]
- 71.Geiler-Samerotte KA, Zhu YO, Goulet BE, Hall DW, Siegal ML. Selection Transforms the Landscape of Genetic Variation Interacting with Hsp90. PLOS Biology. 2016;14(10):e2000465. doi: 10.1371/journal.pbio.2000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calof AL, Kawauchi S, Santos R, Lopez-Burks ME, Hoang MP, Kitzes LM, Lao T, Lechner MS, Hallgrimsson B, Daniel JA, Nussenzweig A, Lander AD. Insights Into Cornelia de Lange Syndrome From the Nipbl-Mutant Mouse. American Journal of Medical Genetics Part A. 2010;152A(7):1633–1633. [Google Scholar]
- 73.Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA. Multiple organ system defects and transcriptional dysregulation in the Nipbl+/− mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5(9):e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santos R, Kawauchi S, Jacobs RE, Lopez-Burks ME, Choi H, Wikenheiser J, Hallgrimsson B, Jamniczky HA, Fraser SE, Lander AD, Calof AL. Conditional Creation and Rescue of Nipbl-Deficiency in Mice Reveals Multiple Determinants of Risk for Congenital Heart Defects. PLoS Biol. 2016;14(9):e2000197. doi: 10.1371/journal.pbio.2000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harmacek L, Watkins-Chow DE, Chen J, Jones KL, Pavan WJ, Salbaum JM, Niswander L. A unique missense allele of BAF155, a core BAF chromatin remodeling complex protein, causes neural tube closure defects in mice. Dev Neurobiol. 2014;74(5):483–97. doi: 10.1002/dneu.22142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Green RM, Fish JL, Young NM, Smith FJ, Roberts B, Dolan K, Choi I, Leach CL, Gordon P, Cheverud JM, Roseman CC, Williams TJ, Marcucio RS, Hallgrimsson B. Developmental nonlinearity drives phenotypic robustness. Nat Commun. 2017;8(1):1970. doi: 10.1038/s41467-017-02037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Green RM, Feng W, Phang T, Fish JL, Li H, Spritz RA, Marcucio RS, Hooper J, Jamniczky H, Hallgrímsson B, Williams T. Tfap2a-dependent changes in mouse facial morphology result in clefting that can be ameliorated by a reduction in Fgf8 gene dosage. Dis Model Mech. 2015;8(1):31–43. doi: 10.1242/dmm.017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hallgrimsson B, Brown JJ, Ford-Hutchinson AF, Sheets HD, Zelditch ML, Jirik FR. The brachymorph mouse and the developmental-genetic basis for canalization and morphological integration. Evol Dev. 2006;8(1):61–73. doi: 10.1111/j.1525-142X.2006.05075.x. [DOI] [PubMed] [Google Scholar]
- 79.Richardson JB, Uppendahl LD, Traficante MK, Levy SF, Siegal ML. Histone Variant HTZ1 Shows Extensive Epistasis with, but Does Not Increase Robustness to, New Mutations. PLOS Genetics. 2013;9(8):e1003733. doi: 10.1371/journal.pgen.1003733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forsberg SK, Andreatta ME, Huang XY, Danku J, Salt DE, Carlborg O. The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance. PLoS Genet. 2015;11(11):e1005648. doi: 10.1371/journal.pgen.1005648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen X, Pettersson M, Rönnegård L, Carlborg Ö. Inheritance Beyond Plain Heritability: Variance-Controlling Genes in Arabidopsis thaliana. PLOS Genetics. 2012;8(8):e1002839. doi: 10.1371/journal.pgen.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilkins JF. Genomic imprinting and methylation: epigenetic canalization and conflict. Trends in Genetics. 2005;21(6):356–365. doi: 10.1016/j.tig.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 83.Pujadas E, Feinberg AP. Regulated noise in the epigenetic landscape of development and disease. Cell. 2012;148(6):1123–31. doi: 10.1016/j.cell.2012.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Faulk C, Barks A, Dolinoy DC. Phylogenetic and DNA methylation analysis reveal novel regions of variable methylation in the mouse IAP class of transposons. BMC Genomics. 2013;14:48–48. doi: 10.1186/1471-2164-14-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morgan HD, Sutherland HGE, Martin DIK, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nature Genetics. 1999;23:314. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 86.Juriloff DM, Harris MJ, Mager DL, Gagnier L. Epigenetic mechanism causes Wnt9b deficiency and nonsyndromic cleft lip and palate in the A/WySn mouse strain. Birth Defects Res A Clin Mol Teratol. 2014;100(10):772–88. doi: 10.1002/bdra.23320. [DOI] [PubMed] [Google Scholar]
- 87.Plamondon JA, Harris MJ, Mager DL, Gagnier L, Juriloff DM. The clf2 gene has an epigenetic role in the multifactorial etiology of cleft lip and palate in the A/WySn mouse strain. Birth Defects Research Part A: Clinical and Molecular Teratology. 2011;91(8):716–727. doi: 10.1002/bdra.20788. [DOI] [PubMed] [Google Scholar]
- 88.Parsons TE, Kristensen E, Hornung L, Diewert VM, Boyd SK, German RZ, Hallgrimsson B. Phenotypic variability and craniofacial dysmorphology: increased shape variance in a mouse model for cleft lip. Journal of Anatomy. 2008;212(2):135–43. doi: 10.1111/j.1469-7580.2007.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nichols JT, Blanco-Sanchez B, Brooks EP, Parthasarathy R, Dowd J, Subramanian A, Nachtrab G, Poss KD, Schilling TF, Kimmel CB. Ligament versus bone cell identity in the zebrafish hyoid skeleton is regulated by mef2ca. Development. 2016;143(23):4430–4440. doi: 10.1242/dev.141036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bosia C, Osella M, Baroudi ME, Cora D, Caselle M. Gene autoregulation via intronic microRNAs and its functions. BMC Syst Biol. 2012;6:131. doi: 10.1186/1752-0509-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515–24. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends in genetics. 2009;25(9):395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318(5848):271–4. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 95.Kasper DM, Moro A, Ristori E, Narayanan A, Hill-Teran G, Fleming E, Moreno-Mateos M, Vejnar CE, Zhang J, Lee D, Gu M, Gerstein M, Giraldez A, Nicoli S. MicroRNAs Establish Uniform Traits during the Architecture of Vertebrate Embryos. Dev Cell. 2017;40(6):552–565 e5. doi: 10.1016/j.devcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cassidy JJ, Jha AR, Posadas DM, Giri R, Venken KJ, Ji J, Jiang H, Bellen HJ, White KP, Carthew RW. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell. 2013;155(7):1556–67. doi: 10.1016/j.cell.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goulart LF, Bettella F, Sønderby IE, Schork AJ, Thompson WK, Mattingsdal M, Steen VM, Zuber V, Wang Y, Dale AM, Andreassen OA, Djurovic S. MicroRNAs enrichment in GWAS of complex human phenotypes. BMC Genomics. 2015;16(1):304. doi: 10.1186/s12864-015-1513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aubin D. Forms of explanation in the catastrophe theory of René Thom: topology, morphogenesis, and the structuralism, Growing explanations: Historical perspectives on recent science. Duke Univ. Press; 2004. pp. 95–130. [Google Scholar]
- 99.Newman SA, Rice SA. Model for Constraint and Control in Biochemical Networks. Proceedings of the National Academy of Sciences. 1971;68(1):92–96. doi: 10.1073/pnas.68.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kauffman S. The large scale structure and dynamics of gene control circuits: An ensemble approach. Journal of Theoretical Biology. 1974;44(1):167–190. doi: 10.1016/s0022-5193(74)80037-8. [DOI] [PubMed] [Google Scholar]
- 101.Xiao Y. A Tutorial on Analysis and Simulation of Boolean Gene Regulatory Network Models. Current Genomics. 2009;10(7):511–525. doi: 10.2174/138920209789208237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Soulé ME. Allomeric variation 1.: The theory and some consequences. The American Naturalist. 1982;120(6):751–764. [Google Scholar]
- 103.Barbaric I, Miller G, Dear TN. Appearances can be deceiving: phenotypes of knockout mice. Briefings in Functional Genomics. 2007;6(2):91–103. doi: 10.1093/bfgp/elm008. [DOI] [PubMed] [Google Scholar]
- 104.Wagner A. Evolution of gene networks by gene duplications: a mathematical model and its implications on genome organization. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(10):4387–4391. doi: 10.1073/pnas.91.10.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morishita Y, Iwasa Y. Accuracy of positional information provided by multiple morphogen gradients with correlated noise. Physical Review E. 2009;79(6):061905. doi: 10.1103/PhysRevE.79.061905. [DOI] [PubMed] [Google Scholar]
- 106.Levy SF, Siegal ML. Network Hubs Buffer Environmental Variation in Saccharomyces cerevisiae. PLOS Biology. 2008;6(11):e264. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Geiler Samerotte K, Sartoria FMO, Siegal ML. Decanalizing thinking on Canalization. Seminars in Cell and Developmental Biology. doi: 10.1016/j.semcdb.2018.05.008. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.White RJ, Nie Q, Lander AD, Schilling TF. Complex Regulation of cyp26a1 Creates a Robust Retinoic Acid Gradient in the Zebrafish Embryo. PLOS Biology. 2007;5(11):e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paulsen M, Legewie S, Eils R, Karaulanov E, Niehrs C. Negative feedback in the bone morphogenetic protein 4 (BMP4) synexpression group governs its dynamic signaling range and canalizes development. Proceedings of the National Academy of Sciences. 2011;108(25):10202–10207. doi: 10.1073/pnas.1100179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nelson RM, Pettersson ME, Li X, Carlborg Ö. Variance Heterogeneity in Saccharomyces cerevisiae Expression Data: Trans-Regulation and Epistasis. PLOS ONE. 2013;8(11):e79507. doi: 10.1371/journal.pone.0079507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.von Dassow M, Davidson LA. Physics and the canalization of morphogenesis: a grand challenge in organismal biology. Phys Biol. 2011;8(4):045002. doi: 10.1088/1478-3975/8/4/045002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lerner IM. Genetic Homeostasis. Wiley & Sons; New York: 1954. [Google Scholar]
- 113.Hartl GB, Suchentrunk F, Willing R, Petznek R. Allozyme heterozygosity and fluctuating asymmetry in the brown hare (L epus europaeus): A test of the developmental homeostasis hypothesis. Philos Trans R Soc Lond [Biol] 1995;350:313–323. [Google Scholar]
- 114.Suchentrunk F. Variability of minor tooth traits and allozymic diversity in brown hare Lepus europaeus populations. Acta Theriologica. 1993;38(Suppl 2):59–69. [Google Scholar]
- 115.Mitton JB. Enzyme heterozygosity, metabolism, and developmental stability. Genetica. 1993;89:47–65. [Google Scholar]
- 116.Livshits G, Kobyliansky E. Fluctuating asymmetry as a possible measure of developmental homeostasis in humans: a review. Human Biology. 1991;63(4):441–466. [PubMed] [Google Scholar]
- 117.Bader RS. Fluctuating Asymmetry in the dentition if the house mouse. Growth. 1965;29:291–300. [PubMed] [Google Scholar]
- 118.Clarke GM. The genetic basis of developmental stability. I. Relationships between stability, heterozygosity and genomic coadaptation. Genetica. 1993;89:15–23. [Google Scholar]
- 119.Alibert P, Renaud S, Dod B, Bonhomme F, Auffray JC. Fluctuating asymmetry in the Mus musculus hybrid zone: a heterotic effect in disrupted co-adapted genomes. Proceedings of the Royal Society of London B. 1994;258(1351):53–9. doi: 10.1098/rspb.1994.0141. [DOI] [PubMed] [Google Scholar]
- 120.Auffray JC, Fontanillas P, Catalan J, Britton-Davidian J. Developmental stability in house mice heterozygous for single Robertsonian fusions. J Hered. 2001;92(1):23–9. doi: 10.1093/jhered/92.1.23. [DOI] [PubMed] [Google Scholar]
- 121.Graham JH. Genomic coadaptation and developmental stability in hybrid zones. Acta Zoologica Fennica. 1992;191:121–131. [Google Scholar]
- 122.Leary RF, Allendorf FW, Knudsen KL, Thorgaard GH. Heterozygosity and developmental stability in gynogenetic diploid and triploid rainbow trout. Heredity (Edinburgh) 1985;54(Pt 2):219–25. doi: 10.1038/hdy.1985.29. [DOI] [PubMed] [Google Scholar]
- 123.Roff D. Evolutionary quantitative genetics: Are we in danger of throwing out the baby with the bathwater? Annales Zoologici Fennici, JSTOR. 2003:315–320. [Google Scholar]
- 124.Magnus P, Berg K, Bjerkedal T. Association of parental consanguinity with decreased birth weight and increased rate of early death and congenital malformations. Clinical Genetics. 1985;28(4):335–342. doi: 10.1111/j.1399-0004.1985.tb00407.x. [DOI] [PubMed] [Google Scholar]
- 125.Bittles AH, Black ML. Consanguinity, human evolution, and complex diseases. Proceedings of the National Academy of Sciences. 2010;107(suppl 1):1779–1786. doi: 10.1073/pnas.0906079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stinchcombe JR, Kirkpatrick M. Genetics and evolution of function-valued traits: understanding environmentally responsive phenotypes. Trends in Ecology & Evolution. 2012;27(11):637–647. doi: 10.1016/j.tree.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 127.Rice S. A general population genetic theory for the evolution of developmental interactions. PNAS. 2002;99(24):15518–15523. doi: 10.1073/pnas.202620999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morrissey MB. Evolutionary quantitative genetics of nonlinear developmental systems. Evolution. 2015;69(8):2050–66. doi: 10.1111/evo.12728. [DOI] [PubMed] [Google Scholar]
- 129.Klingenberg CP, Nijhout HF. Genetics of fluctuating asymmetry: A developmental model of developmental instability. Evolution. 1999;53(2):358–375. doi: 10.1111/j.1558-5646.1999.tb03772.x. [DOI] [PubMed] [Google Scholar]
- 130.Ramler D, Mitteroecker P, Shama LN, Wegner KM, Ahnelt H. Nonlinear effects of temperature on body form and developmental canalization in the threespine stickleback. J Evol Biol. 2014 doi: 10.1111/jeb.12311. [DOI] [PubMed] [Google Scholar]
- 131.Hartl DL, Dykhuizen DE, Dean AM. Limits of adaptation: the evolution of selective neutrality. Genetics. 1985;111(3):655–74. doi: 10.1093/genetics/111.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wright S. Evolution and the Genetics of Populations, Volume 3: Experimental results and evolutionary deductions. University of Chicago Press; Chicago: 1977. [Google Scholar]
- 133.Badyaev AV, Foresman KR. Extreme environmental change and evolution: stress-induced morphological variation is strongly concordant with patterns of evolutionary divergence in shrew mandibles. Proc R Soc Lond B Biol Sci. 2000;267(1441):371–7. doi: 10.1098/rspb.2000.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Steiner UK, van Buskirk J. Environmental stress and the costs of whole-organism phenotypic plasticity in tadpoles. J Evol Biol. 2008;21(1):97–103. doi: 10.1111/j.1420-9101.2007.01463.x. [DOI] [PubMed] [Google Scholar]
- 135.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol. 1910;40:4–7. [Google Scholar]
- 136.Weiss JN. The Hill equation revisited: uses and misuses. The FASEB Journal. 1997;11(11):835–841. [PubMed] [Google Scholar]
- 137.Steinacher A, Bates DG, Akman OE, Soyer OS. Nonlinear Dynamics in Gene Regulation Promote Robustness and Evolvability of Gene Expression Levels. PLOS ONE. 2016;11(4):e0153295. doi: 10.1371/journal.pone.0153295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zarai Y, Margaliot M, Tuller T. Optimal Down Regulation of mRNA Translation. Scientific Reports. 2017;7:41243. doi: 10.1038/srep41243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lander AD, Nie Q, Wan FY. Do morphogen gradients arise by diffusion? Developmental cell. 2002;2(6):785–796. doi: 10.1016/s1534-5807(02)00179-x. [DOI] [PubMed] [Google Scholar]
- 140.Zhang YT, Alber MS, Newman SA. Mathematical modeling of vertebrate limb development. Mathematical biosciences. 2013;243(1):1–17. doi: 10.1016/j.mbs.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 141.Le Rouzic A, Alvarez-Castro JM, Hansen TF. The Evolution of Canalization and Evolvability in Stable and Fluctuating Environments. Evolutionary Biology. 2013;40(3):317–340. [Google Scholar]
- 142.Pelabon C, Hansen TF, Carter AJ, Houle D. Evolution of variation and variability under fluctuating, stabilizing, and disruptive selection. Evolution. 2010;64(7):1912–25. doi: 10.1111/j.1558-5646.2010.00979.x. [DOI] [PubMed] [Google Scholar]
- 143.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–4. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 144.Huizinga J, Stanley KO, Clune J. The Emergence of Canalization and Evolvability in an Open-Ended, Interactive Evolutionary System. 2017 doi: 10.1162/artl_a_00263. arXiv preprint arXiv:1704.05143. [DOI] [PubMed] [Google Scholar]
- 145.Kim MS, Kim JR, Kim D, Lander AD, Cho KH. Spatiotemporal network motif reveals the biological traits of developmental gene regulatory networks in Drosophila melanogaster. BMC Systems Biology. 2012;6(1):31. doi: 10.1186/1752-0509-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Young NM, Chong HJ, Hu D, Hallgrímsson B, Marcucio RS. Quantitative analyses link modulation of sonic hedgehog signaling to continuous variation in facial growth and shape. Development. 2010;137(20):3405–3409. doi: 10.1242/dev.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lewontin RC. The genetic basis of evolutionary change. Columbia University Press; New York: 1974. [Google Scholar]
- 148.Xu Q, Jamniczky H, Hu D, Green RM, Marcucio RS, Hallgrimsson B, Mio W. Correlations between the morphology of sonic hedgehog expression domains and embryonic craniofacial shape. Evolutionary biology. 2015;42(3):379–386. doi: 10.1007/s11692-015-9321-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Epp JR, Niibori Y, Liz Hsiang HL, Mercaldo V, Deisseroth K, Josselyn SA, Frankland PW. Optimization of CLARITY for Clearing Whole-Brain and Other Intact Organs(1,2,3) eNeuro. 2015;2(3) doi: 10.1523/ENEURO.0022-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Martinez-Abadias N, Mateu Estivill R, Sastre Tomas J, Motch Perrine S, Yoon M, Robert-Moreno A, Swoger J, Russo L, Kawasaki K, Richtsmeier J, Sharpe J. Quantification of gene expression patterns to reveal the origins of abnormal morphogenesis. bioRxiv. 2018 doi: 10.7554/eLife.36405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Martínez-Abadías N, Mateu R, Niksic M, Russo L, Sharpe J. Geometric Morphometrics on Gene Expression Patterns Within Phenotypes: A Case Example on Limb Development. Systematic Biology. 2016;65(2):194–211. doi: 10.1093/sysbio/syv067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hu D, Young NM, Xu Q, Jamniczky H, Green RM, Mio W, Marcucio RS, Hallgrimsson B. Signals from the brain induce variation in avian facial shape. Dev Dyn. 2015;244(9):1133–1143. doi: 10.1002/dvdy.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Matamoro-Vidal A, Salazar-Ciudad I, Houle D. Making quantitative morphological variation from basic developmental processes: Where are we? The case of the Drosophila wing. Developmental Dynamics. 2015;244(9):1058–1073. doi: 10.1002/dvdy.24255. [DOI] [PMC free article] [PubMed] [Google Scholar]