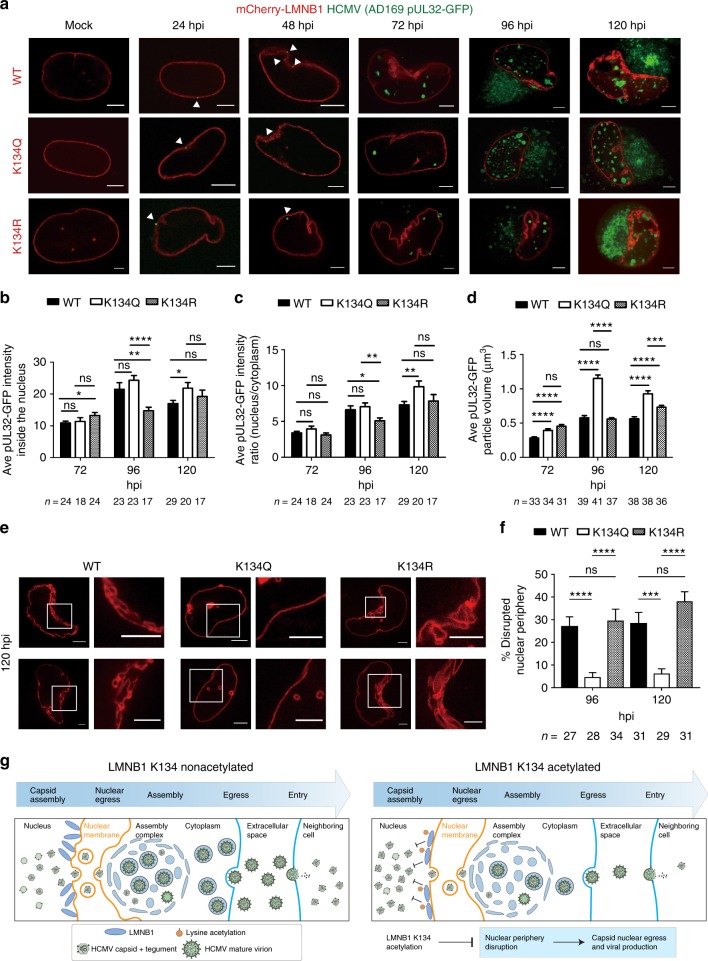
Fig. 6.
LMNB1 acetylation impedes lamina disruption and viral capsid nuclear egress. a Live cell confocal fluorescence microscopy of infection time course in cells transfected with WT, K134Q, or K134R mCherry-LMNB1 (red) and infected with HCMV AD169 UL32-GFP (green). One slice through the center of the nucleus is shown. Representative images are shown; scale bar = 5 μm. b, c pUL32-GFP nuclear intensity in WT, K134Q, or K134R at each time point; the number of cells (n) is indicated. A two-sided Student’s t test was used. b Mean pUL32-GFP intensity across the center three slices inside the nucleus + SEM. c The ratio of mean pUL32-GFP intensity inside the nucleus to outside the nucleus across the center three slices + SEM. d The volume of the pUL32-GFP particles was analyzed in a 3D rending of each Z-stack, and the number of analyzed cells (n) is indicated. The mean pUL32-GFP particle volume + SEM is shown. A two-sided Student’s t test was used. *p value ≤ 0.05, **p value ≤ 0.01, ***p value ≤ 0.001, ****p value ≤ 0.0001. e Live cell confocal fluorescence microscopy of nuclei at 120 hpi for cells transfected with WT, K134Q, or K134R mCherry-LMNB1 (red) constructs. Left: one representative slice through the center of the nucleus. Right: zoom-in of the square on the left; scale bar = 5 μm. f Percentage of nuclear periphery disruption. Disruption in nuclear periphery are seen as infoldings, as visualized by mCherry-LMNB1 fluorescence across center slices. Mean percentage disruption + SEM is shown and cell numbers are indicated. A two-sided Student’s t test was used; ***p value ≤ 0.001, ****p value ≤ 0.0001. g Proposed model for the function of LMNB1 K134 acetylation. Acetylation may stabilize LMNB1, which may inhibit nuclear periphery deformation and thereby impede HCMV capsid nuclear egress and infectious virus production. Source data are provided as a Source Data file