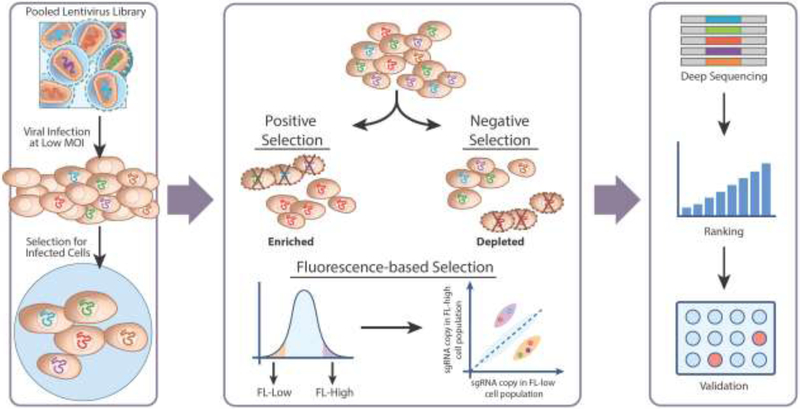
Figure 1. Pooled CRISPR screening strategies.
In a pooled CRISPR screen, a lentiviral sgRNA library is transduced into a cell population at a low multiplicity of infection (MOI) with the goal that each cell receives only one sgRNA integration. Infected cells are put through a positive or negative selection process (i.e. drugs, virus infections, etc.) to identify sgRNAs that are either enriched or depleted, respectively, in the surviving cells through next-generation sequencing. One may also design the screen based on the effect of the sgRNA targeting on gene expression using FACS based on a fluorescent reporter or antibody staining: the relative sgRNA abundance in fluorescence-low (FL-low) versus fluorescence-high (FL-high) cell populations can be quantified. The sgRNAs and corresponding genes are then ranked based on the degree of sgRNA depletion or enrichment, and potential candidate gene hits can then be further validated.