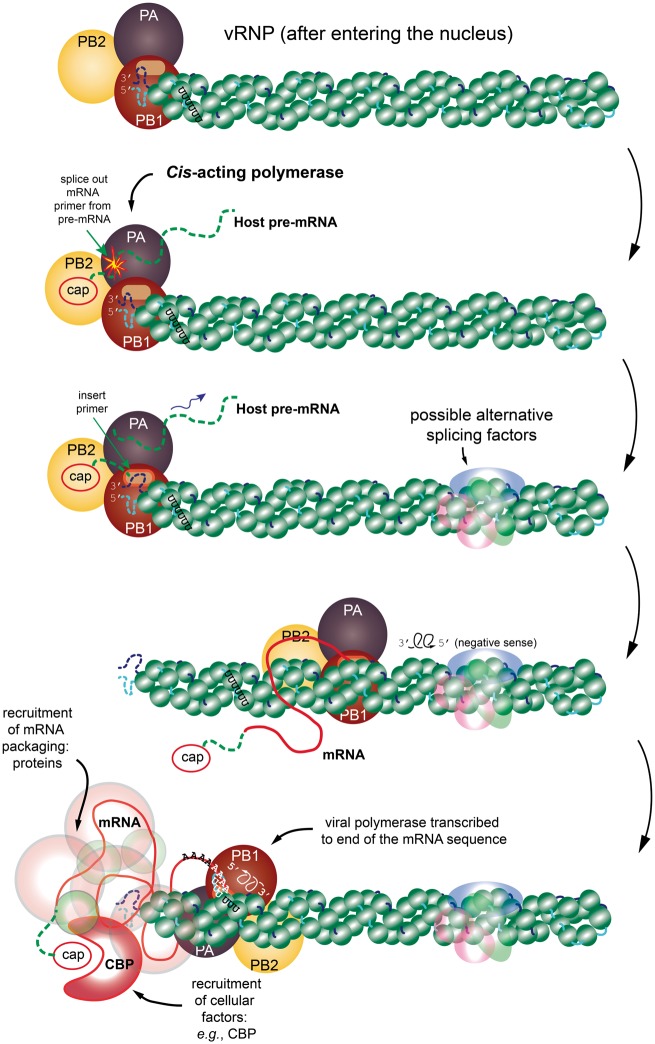
Figure 1.
A cartoon representation of the transcription process. The figure shows the time-dependent and structural relationships of the vRNP and associated viral polymerase, which consists of the subunits PB1, PB2 and PA (from genome Segments 2, 1 and 3, respectively). The viral polymerase binds to the partially complementary 5′ and 3′ termini of the vRNA segment. The progression is from top to bottom. (Top) The vRNP has entered the nucleus of the host cell and has separated from other vRNPs. (Next step) The transcription is initiated by PB2 hijacking a 5′ cap from the host pre-mRNA, followed by endonucleolytic cleavage of the host pre-mRNA by PA. (Next frame) Transcription begins from the 3′ end of the vRNA sequence with the spliced-in primer sequence and cap merged onto the forming mRNA sequence. (Next frame) Transcription continues with the viral polymerase somehow dislodging the NP in its wake to obtain the mRNA transcript. Also depicted faintly are alternative splice factors from the spliceosome that can influence the resulting transcription. (Next frame) While the transcription process is happening, the vRNA and NP tend to recombine back to the rest state. (Final step) The poly-adenylation step is reached where further editing is required of the structure using the host RNA polymerase II system. It is important to remember that during this entire process, various mRNA packaging proteins are being recruited (e.g. the CBP. These are shown as a faint covering of the final mRNA. All this packaging is required for transport to the cytosol for translation into viral proteins. The schematic is inspired from [44] and [42]. The structure of the vRNP is based on the right-handed helix in [42]; however, it is important to remember that [43] reports a left-handed helix. (A colour version of this figure is available online at: https://academic.oup.com/bfg)