 Plants are sessile organisms. To compensate for not being able to escape when challenged by unfavorable growth conditions, pests or herbivores, plants have perfected their metabolic plasticity by having developed the capacity for on demand dynamic biosynthesis and storage of a plethora of phytochemicals.
Plants are sessile organisms. To compensate for not being able to escape when challenged by unfavorable growth conditions, pests or herbivores, plants have perfected their metabolic plasticity by having developed the capacity for on demand dynamic biosynthesis and storage of a plethora of phytochemicals.
Abstract
Covering: up to 2018
Plants are sessile organisms. To compensate for not being able to escape when challenged by unfavorable growth conditions, pests or herbivores, plants have perfected their metabolic plasticity by having developed the capacity for on demand synthesis of a plethora of phytochemicals to specifically respond to the challenges arising during plant ontogeny. Key steps in the biosynthesis of phytochemicals are catalyzed by membrane-bound cytochrome P450 enzymes which in plants constitute a superfamily. In planta, the P450s may be organized in dynamic enzyme clusters (metabolons) and the genes encoding the P450s and other enzymes in a specific pathway may be clustered. Metabolon formation facilitates transfer of substrates between sequential enzymes and therefore enables the plant to channel the flux of general metabolites towards biosynthesis of specific phytochemicals. In the plant cell, compartmentalization of the operation of specific biosynthetic pathways in specialized plastids serves to avoid undesired metabolic cross-talk and offers distinct storage sites for molar concentrations of specific phytochemicals. Liquid–liquid phase separation may lead to formation of dense biomolecular condensates within the cytoplasm or vacuole allowing swift activation of the stored phytochemicals as required upon pest or herbivore attack. The molecular grid behind plant plasticity offers an endless reservoir of functional modules, which may be utilized as a synthetic biology tool-box for engineering of novel biological systems based on rational design principles. In this review, we highlight some of the concepts used by plants to coordinate biosynthesis and storage of phytochemicals.
1. Introduction
Phytochemicals play a key role in plant defense, communication and adaptation to abiotic and biotic stress. Derived from relatively few general metabolites, phytochemicals diversify into an immense number of molecules through combinatorial enzyme cascade reactions.1 With enzymatic precision, plants produce complex phytochemicals with regio- and stereospecific functional groups matching or outperforming modern day synthetic chemistry.2 Their biosynthesis often involves assemblies of numerous enzymes, many of which appear to have low substrate affinity and specificity when studied in vitro. However, in planta the metabolism may be highly channeled3,4 and tends to work more efficient than would be expected from the average cellular concentrations of the biosynthetic enzymes and their substrates.5,6 These serial enzyme reactions require tight regulation to guide the metabolism in a highly crowded environment.7,8
In 1978 Charles Tanford wrote: “Biological organization may be viewed as consisting of two stages: biosynthesis and assembly”.9 In plants, this is achieved through spatial organization of biosynthetic enzymes and metabolites in compartments dictating metabolic flux into streamlined coordinated highways.10,11 The classical view of compartments as clearly separated entities confined by membranes is being challenged with the advances of analytical tools with subcellular resolution. Instead, a dynamic intracellular web of the endoplasmic reticulum (ER) interacting with cellular compartments and formation of local microcompartments governs the plasticity of plant metabolism.12 Compartmentalization at the nanoscale level by organization of enzymes in confined spaces at the membrane surfaces13 or in membrane-less assemblies obtained by liquid–liquid phase separation (LLPS)14 provides dynamic assemblies necessary for metabolic adaptation.
In this review, we focus on new insights on plant compartmentalization and the dynamic solutions plants use to cope with their sessile life style. We also discuss how compartmentalization may be used as a synthetic biology tool to optimize the production of phytochemicals in heterologous hosts.
2. Dynamic assembly of biosynthetic enzyme complexes
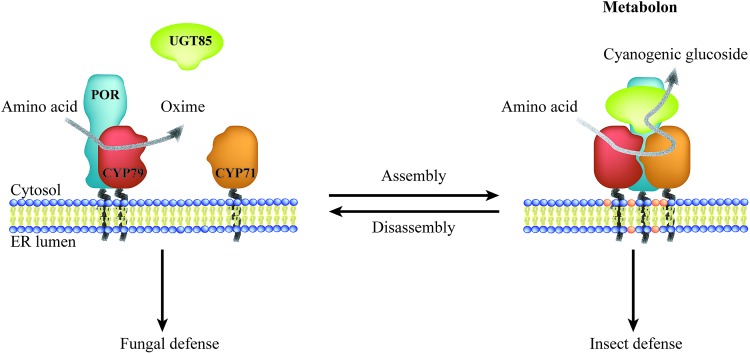
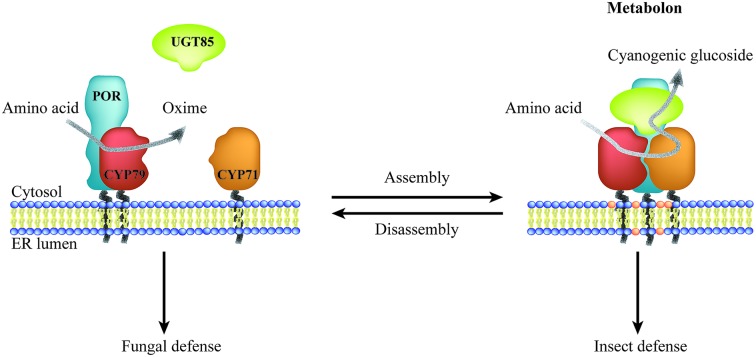
Biosynthesis of phytochemicals typically involves multiple enzymatic steps, and spatial confinement of biosynthetic enzymes governs the formation of metabolic highways. Most phytochemicals are products of biosynthetic pathways, in which enzymes of the cytochrome P450 (P450) superfamily catalyze one or more key steps.15 Microsomal P450 enzymes are tethered to the ER via a single transmembrane anchor and thereby confined to the two-dimensional membrane lattice. Stoichiometric imbalances between P450s and their oxidoreductase, NADPH-dependent cytochrome P450 oxidoreductase (POR), ranges between 6 : 1 to 20 : 1,6,16–19 and the overall low abundance suggest that these enzymes possess features guiding them towards specific preferential interactions thus circumventing bulk equilibrium.20–22 Metabolic channeling within biosynthetic pathways may be achieved through organization of enzymes in complexes, termed metabolons.23 Efficient channeling requires close proximity (0.1–1 nm) between sequential enzymes24 as achieved by protein–protein interactions. Consequently, metabolon assembly facilitates direct transfer of substrates and products between sequential enzymes and prevents leakage of potentially toxic and labile intermediates and undesired metabolic cross talk. Dynamic assembly and disassembly of metabolons offer an opportunity for swift adaption to meet environmental challenges such as fungal or insect attack (Fig. 1). The on-demand organization of POR with specific P450s would surpass the challenge of the stoichiometric imbalance between the enzymes.
Fig. 1. Illustration of the dynamic assembly and disassembly of the dhurrin metabolon. Metabolon assembly involves recruitment of the soluble UGT to the ER-anchored P450s and POR and local accumulation of specific lipids resulting in biosynthesis of the insecticide dhurrin. Disassembly of the dhurrin metabolon results in release of the antifungal oxime intermediate. Thus, a single pathway confers resistance towards specific herbivores and pests governed by the dynamic assembly of enzymes in metabolons.
2.1. Metabolons; the highways of plant metabolism
Biosynthetic pathways for phytochemical production in the plant cell can be highly branched sharing a few common steps. Assembly of sequential enzymes in metabolons provides a way of orchestrating the metabolic grid by guiding the metabolites towards a specific product. One key example is the phenylpropanoid pathway, which directs the production towards monolignols used in lignin biosynthesis or towards bioactive flavonoids depending on the plant's need.6,25–27 The latter pathway is further differentially divided into sub-branches with different end products such as vanilloids, isoflavonoids, flavonols, flavones, anthocyanins etc.
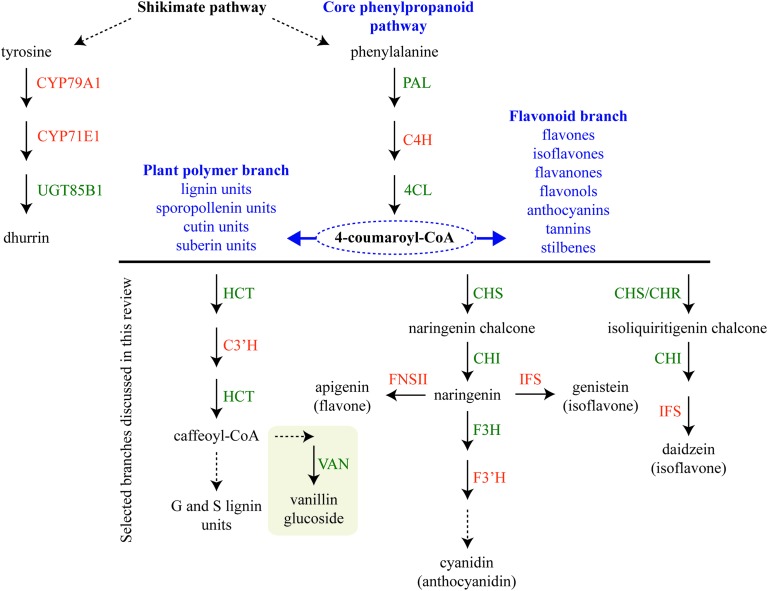
Assembly of metabolons facilitate channeling of phenylalanine through the core phenylpropanoid pathway28 and further downstream towards the monolignols,29 flavonoids,25,26 sporopollenin branches27,30 and vanilloids such as vanillin glucoside (Fig. 2). Formation of enzymatic complexes in the phenylpropanoid pathway was first proposed in 1974.31 Early research supported this hypothesis based on substrate channeling and interactions between two enzymes in the core phenylpropanoid pathway, cinnamate-4-hydroxylase (C4H, CYP73A) and phenylalanine ammonia-lyase (PAL)32 (Fig. 2) and interaction between soluble members in downstream branches, for reviews on this topic see ref. 4, 5, 33 and 34.
Fig. 2. Schematic representation of dhurrin biosynthesis (left) derived from the aromatic amino acid tyrosine and phenylpropanoid biosynthesis derived from the aromatic amino acid phenylalanine. The core phenylpropanoid pathway branches out into a metabolic grid of downstream pathways leading to the production of e.g. flavonoids and stilbenes and monomeric units as building blocks for plant polymers. Selected phenylpropanoid branches discussed in the main text are presented. P450 enzymes are colored in orange and other enzymes are colored in green. Abbreviations: C3′H, p-coumaroylshikimate 3′-hydroxylase (CYP98A); C4H, cinnamate-4-hydroxylase (CYP73A); CHI, chalcone isomerase; CHR, chalcone reductase; CHS, chalcone synthase, 4CL, 4-coumaric acid CoA ligase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′ hydroxylase (CYP75B); FNSII, flavone synthase II (CYP93B); HCT, hydroxycinnamoyl transferase; IFS, isoflavone synthase; PAL, phenylalanine lyase; VAN, vanillin synthase. Broken arrows indicate multistep enzymatic reactions. Box around vanillin glucoside indicates compartmentalization.
The mechanisms governing the assembly of metabolons remains largely elusive. However, recent publications support the role of P450s to recruit soluble pathway partners and to serve as nucleation sites for metabolon assembly.6,25,26,29,35 For example, co-expression of the arabidopsis (Arabidopsis thaliana) lignin biosynthetic enzyme p-coumaroylshikimate 3′-hydroxylase (AtC3′H, CYP83A3) with hydroxycinnamoyl transferase (AtHCT) or 4-coumaric acid CoA ligase (At4-CL) in Nicotiana benthamiana shifted localization of the soluble AtHCT and At4-CL towards the ER.29 Localization of the soluble enzymes was less effected by AtC4H (CYP73A5), however, association between AtC4H and AtHCT was increased when AtC3′H and At4-CL were co-expressed indicating that the presence of more downstream partners stimulates the assembly of the core phenylpropanoid pathway enzymes. In isoflavonoid biosynthesis in soybean (Glycine max), chalcone isomerase (GmCHI) and chalcone synthase (GmCHS) were shown to interact with the isoflavone synthase (GmIFS, CYP93C) on the surface of the ER.25,36 Chalcone reductase (GmCHR) only interacted with one of the two IFS homologs25 pointing to isoform preference as a controlling factor for metabolon formation. In the branch leading to flavones in the metabolic grid of snapdragon (Antirrhinum majus) flavonoids, AmCHI and AmCHS were found to interact with flavone synthase II (AmFNSII, CYP93B) on the ER. In the competing branch for anthocyanin production, flavonoid 3′-hydroxylase (AmF3′H, CYP75B) was also found to interact with the upstream AmCHI.26 During flower development, accumulation of flavones takes off before accumulation of anthocyanins in petals and thereafter both flavonoid classes accumulate.26 How the flavonoid precursors are differentially divided into the two co-occurring branches remains elusive. Yet, it is likely that formation of metabolons plays a key role in guiding and controlling the phytochemical profile.
Assembly of biosynthetic pathways into metabolons is not restricted to the phenylpropanoids. Metabolon formation of the biosynthetic enzymes catalyzing production of the defense compound dhurrin in sorghum (Sorghum bicolor) has been studied for decades. In sorghum, dhurrin is synthesized from l-tyrosine by the sequential action of two P450s (CYP79A1 and CYP71E1), POR20–22,37–40 and a soluble UDP-glucose dependent glucosyltransferase (UGT85B1)41–48 (Fig. 1). Early studies in the 1980s using sorghum microsomes showed tight channeling of intermediates in dhurrin biosynthesis.3,49 The pathway has been studied intensively since then and in 2016 isolation of the dhurrin metabolon was reported together with functional and structural characteristics of the complex.6 Finally, the role of P450s in the assembly of metabolons has also been implied in the biosynthesis of carotenoids.50
2.2. Organization and dynamics of metabolons
The dynamic nature of transient metabolons makes it difficult to document their existence as illustrated by the many years of research on metabolon formation in the biosynthesis of phenylpropanoids and cyanogenic glucosides e.g. dhurrin. Much research on metabolons, especially in the metabolic grid of phenylpropanoids, has focused mainly on co-localization and binary interactions between enzymes. This has been studied using experimental systems such as the yeast-two-hybrid or split-ubiquitin versions, bimolecular fluorescence complementation (BiFC), fluorescence (Förster) resonance energy transfer (FRET) and co-immunoprecipitation (Co-IP). The latter is sometimes combined with proteomics to investigate additional interaction partners. Less is known about factors stimulating assembly of enzymes, flow of metabolites and organization of these dynamic complexes with different configurations. Some insight on the structural organization and regulation of metabolons has transpired in the recent years and future advances in molecular technologies will allow for more detailed knowledge on this subject.
Advances in FRET-based techniques and instrumentation provide more robust data on protein–protein interactions by reducing the number of false positives51 and provide comprehensive knowledge about the organization of the proteins under investigation. Organization of the dhurrin biosynthetic enzymes in planta has been studied using fluorescence correlation spectroscopy (FCS) and FRET upon transient expression in N. benthamiana.6 CYP79A1, CYP71E1 and UGT85B1 were found to form Homo- and hetero-oligomerization, with an enhancement of UGT85B1 oligomerization when CYP79A1 or CYP71E1 was co-expressed. Homo- and hetero-oligomers of P450s is also known from the lignin pathway in arabidopsis and poplar (Populus trichocarpa),29,52 and might be a more general feature of P450-containing metabolons.
Assembly of sequential enzymes in metabolons serves to guide the direction and flux of metabolites. Interaction with downstream soluble pathway partners may have a positive effect on enzyme catalysis. In the dhurrin metabolon, recruitment of the soluble UGT85B1 to the P450s stimulated the channeling between CYP79A1 and CYP71E1, even in the absence of its UDP-glucose cofactor.6 Interaction between P450s may also promote catalytic activity. Biosynthesis of the carotenoid lutein in Escherichia coli catalyzed by CYP97C required co-expression of CYP97A, indicating a synergistic interaction between the two P450s, which are chloroplast-localized in plants.53
Metabolons are stabilized by weak protein–protein interactions, which has prevented their isolation by use of classical detergents because application of such detergents results in dissociation of the enzyme complexes. Alternatives to detergents hold promise for isolation of metabolons. A co-polymer of styrene and maleic acid (SMA), which spontaneously inserts into membranes and form discrete lipid particles (SMALPs), was recently applied to isolate membrane-bound constituents of the dhurrin metabolon.6 SMALPs were prepared from sorghum microsomes and subjected to affinity purification using POR as bait resulting in an enrichment of both CYP79A1 and CYP71E1 in purified samples. The SMALP platform, in combination with state-of-the-art methods such as single particle cryo-electron microscopy (EM), are promising for obtaining structural insight on the organization of metabolons. Cryo-EM has shown potential to provide new structural insights of challenging biological systems and the different configurations there might exist with Ångström scale resolution.54 Cryo-EM has recently been combined with the SMALP technology to study membrane proteins.55–57
The local lipid composition also plays a role in channeling and possibly stimulating assembly of membrane-bound metabolon components such as P450s. Investigation of the lipid composition of the SMALP purified dhurrin metabolons showed an enrichment in lipids with the negatively charged PG head-groups compared to the total sorghum microsomal lipid extract and intriguingly CYP71E1 was shown to exhibit higher catalytic activity in liposomes with 20–30% negatively charged lipids.6 This highlights the importance of studying biosynthetic metabolons in a native-like lipid environment to further investigate the presence of essential lipids associated with membrane-bound metabolons. Advances in native mass spectrometry have made it possible to analyze intact membrane-bound protein complexes together with their associated lipids.58,59 This would enable direct assessment of specific lipids associated with metabolons.
Other structural components of the ER membrane may also influence the organization of membrane-bound metabolon members. In arabidopsis, specific membrane steroid-binding proteins (MSBPs) interact with the phenylpropanoid P450 enzymes AtC4H, AtC3′H and ferulate/coniferaldehyde 5-hydroxylase (AtF5H, CYP84A1).60 The MSBPs were suggested to organize and stabilize the P450s by serving as scaffolds for P450 clustering and thereby be a controlling factor in guiding the metabolism towards biosynthesis of monolignols. Noteworthy, MSBP proteins were also enriched in the purified SMALPs from sorghum microsomes,6 but their possible importance for the assembly of the dhurrin metabolon has not yet been investigated. This illustrates how isolation of native metabolons using the SMA based technology may disclose involvement of previously unrecognized regulatory or structural components.
2.3. Membrane dynamics shape phytochemical production
Intracellular compartments confined by a membrane offer specialized sites for biosynthesis and storage of cell constituents. The formation of compartments is a dynamic process driven by self-assembly of biomolecules that, guided by thermodynamics, fold or assemble into their lowest energetic state. Membranes are composed of lipids that self-assemble to form a two-dimensional lattice stabilized by hydrophobic forces. Theoretical calculations predict that entropic and enthalpic effects influence the energy states during the transition from monomeric lipids to bilayers, respectively.61 This means that clusters larger than 1 nm may be stabilized as the dissolution entropic effects become weaker with increasing particle volume. Cluster formation is concentration dependent, which indicates that simply bringing molecules together may lead to demixing or LLPS and formation of micro-domains. These effects facilitate phase separation of cholesterol, sphingolipids and proteins within the lipid bilayers and formation of functional microcompartments, lipid rafts.62 Some mammalian P450 enzymes have been proposed to organize in lipid raft-like structures.63 It remains uncertain whether P450 enzymes involved in biosynthesis of phytochemicals are organized in a similar fashion.
In addition to the confining role, the membrane serves specialized transport functions, including the import and sorting of proteins and exchange of metabolites. Thus, membrane barriers prevents undesired metabolic cross-talk. Nevertheless, dynamic interplay between compartments is required to provide the metabolic plasticity necessary to enable the plant to adapt to environmental changes. For example, the C5 isoprene precursors of mono- and diterpenoids are produced in the chloroplasts through the MEP pathway,64,65 whereas decorations and functionalization is carried out at the cytosolic surface of the ER membrane by the action of membrane-anchored P450 enzymes66 and cytosolic transferases. These processes may be connected through dynamic ER networks bridging different compartments through specific protein interaction sites.12 Each compartment presents distinct membrane contact sites with high specificity to bring the membranes in close proximity and enable exchange of metabolites and proteins.67,68 Accordingly, evolutionary distinct pathways are stitched together by the ER giving rise to the incredible chemical diversity found in plants. Thus, understanding the dynamics of the ER, interactions with other compartments and formation of microcompartments would enable further exploitation of the plant cell in production of phytochemicals.
3. Classical compartments and phytochemical dynamics
Besides formation of metabolic enzyme complexes, biosynthesis and storage of a vast range of phytochemicals involves dynamic interplay between multiple subcellular compartments and cell types.69,70 The involvement of diverse compartments in phytochemical biosynthesis dictates that intracellular translocation must be highly regulated. Recent technological developments in cell fractionation, protein tagging, immuno-histochemical analyses, advanced microscopic techniques and mass spectrometry imaging (MSI) have shed light on the intracellular compartmentalization of biosynthesis and storage of phytochemicals. Here, we highlight how classical compartments are associated with dynamic phytochemical functions.
As the storage site of the cell's hereditary material, the nucleus plays a key role in governing various cell functions including the coordination of general and specialized metabolism. The orchestration of the biosynthesis of phytochemicals proceeds already at the chromosome level.71 Genes encoding enzymes catalyzing the biosynthesis of several phytochemicals have recently been shown to localize within specific gene clusters. These include different compound classes such as di- and tri-terpenoids (including saponins and steroidal alkaloids), benzylisoquinoline alkaloids, cyanogenic glucosides and benzoxazinoids.71–76 Specific examples of complex biosynthetic pathways encoded by biosynthetic gene clusters are those for the alkaloids thebaine and noscapine in opium poppy (Papaver somniferum). Thebaine, a pentacyclic alkaloid readily converted to the narcotic analgesics codeine and morphine, share a common intermediate S-reticuline with the non-addictive benzylisoquinoline alkaloid noscapine. The thebaine gene cluster is relatively simple encoding five key biosynthetic enzymes, whereas the larger noscapine gene cluster comprises ten genes encoding five distinct enzyme classes (including several P450s).74,77 Following transcription and translation, nuclear encoded enzymes are often targeted to the desired compartment e.g. plastid localized enzymes encoded by nuclear genes.78,79 This targeting and sorting of proteins are governed by the ER, which through contact sites connects all areas of the cell including the plastids and mitochondria.12,68 Although the mitochondria are ‘the power houses’ of the plant cell providing chemical energy in the form of ATP and NADH, and serve vital functions in the biosynthesis and storage of phytochemicals in plants,80,81 in this review we focus on the importance of the plastids in relation to phytochemical biosynthesis and storage as new knowledge has emerged.
3.1. Plastids for production and storage of phytochemicals
Plastids are truly dynamic compartments that differentiate in response to stimuli and cellular metabolism. Chloroplasts are the light-driven power unit of the plant cell catalyzing photosynthetic conversion of CO2 and solar energy to chemical energy. The plant chloroplast is a large ellipsoid compartment (about 5 × 10 μm) confined from the cytosol by an outer and inner envelope and harboring an intricate thylakoid lamellar system. The outer envelope contains porins and is therefore freely permeable for small molecules while the inner envelope is impermeable for ions and metabolites and thereby restricts the free passage of molecules between the cytosol and the interior.82 The chloroplasts have a relatively small genome encoding 80–100 proteins. Accordingly, the vast number of the 2500–3500 chloroplast localized proteins are nuclear encoded and translocated into the compartment.83 In general, nuclear encoded chloroplast proteins are synthesized as precursor proteins with cleavable N-terminal transit peptide that directs each protein to its final destination within the chloroplast sub-compartments.84,85 The chloroplast harbors enzyme systems catalyzing biosynthesis of a remarkably diverse number of both general metabolites and phytochemicals e.g. amino acids,86 fatty acids,87 lipids,88 plant hormones,89 vitamins,90 chlorophyll,91 major diterpenes such as manoyl oxide, miltiradiene, ent-kaurene, vitexifolin92–95 and the chloroplast-bound isoprenoids (β-carotene, lutein, prenyl chains of chlorophylls and plastoquinone-9). Most of these metabolites are vital for the general metabolism of the chloroplast and at different stages of plant development where metabolites are released as signaling components for plant growth and development or as defense compounds against pathogens or herbivores.83
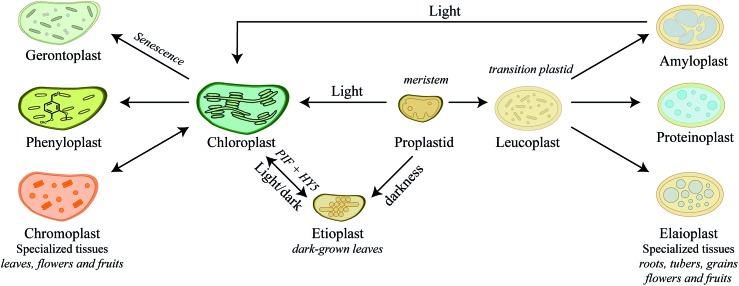
Chloroplasts are by far the most extensively studied plant intracellular compartment due to its vital functionality, yet chloroplast is only one out of a plethora of plastids generated by undifferentiated small proplastids (about 1 μm in diameter) harboring only a few internal membranes and found in meristematic cells.82 The various forms of plastids are interconvertible into each other depending on cell and tissue type, plant growth, development stage, and environmental conditions. The numerous plant plastids each perform specialized functions, such as photosynthesis, biosynthesis of amino acids and fatty acids, storage of carbohydrates and lipids or biosynthesis and storage of phytochemicals and thus play essential roles in plant adaptation. The final number of plastids within a cell and regulatory mechanisms behind plastid biogenesis are far from being understood.96 Here we try to give a short introduction to the biogenesis of plastids and their specialized functions in relation to biosynthesis of phytochemicals (Fig. 3), which has recently been thoroughly reviewed.97–99
Fig. 3. Diversity and differentiation of plastids depending on tissue type and developmental stages. Arrows indicate transitions between different plastid types. The proplastid is the progenitor plastid, point of differentiation, developing into either etioplast or chloroplast depending on exposure to darkness or light, respectively. Proplastids are also able to develop into the intermediate plastid, the leucoplasts that may further differentiate into either amylo-, proteino- or elaioplasts accumulating starch, proteins or oils, respectively. Chloroplasts can also further differentiate depending on environmental stimuli and plant cell types into chromoplasts, phenyloplasts and the dying gerontoplast, in which resources are recycled and redistributed. Chromoplast and phenyloplast accumulate phytochemicals such as carotenoids and phenylpropanoids, respectively.
In meristematic cells, the proplastids are ready to differentiate into the different plastids depending on the developmental stage. Biosynthesis and storage of certain phytochemicals such as carotenoids play a key role in this event. In darkness, proplastids develop into etioplasts (Fig. 3). Upon exposure to light, expression of carotenoid biosynthetic genes, PIFs and HY5, results in a dramatic increase in carotenoid production protecting the plastids from photo-oxidative damage during de-etiolation of etioplasts100 and differentiation of etioplasts into chloroplasts. This process is reversed by longer exposures to darkness. Carotenoids are tetraterpenoids found in most plastids and play vital role in photoprotection throughout plant development.101 In fleshy fruits such as tomatoes, ripening is associated with differentiation of green fruit chloroplasts into ripe fruit chromoplasts102–104 (Fig. 3). Chromoplasts are recognized by massive accumulation of carotenoid pigments that give the red, orange and yellow colors to the plant structure. Upon maturity, the concentration of carotenoids, stored in the lipid-rich microcompartment called plastoglobuli, increases and may result in the formation of carotenoid crystals.104–107 Some plant tissues such as seeds, tubers and leaves contain other specialized plastids that function predominantly as storage sites. As a main carbohydrate energy storage site, amyloplasts prioritize the strong influx of carbon towards biosynthesis of starch compared to carotenoids in chromoplasts. Both chromoplasts and amyloplasts can reversibly differentiate into photosynthetic chloroplasts97–99 as exemplified by the greening of light exposed potato tubers (Fig. 3). In addition to starch storage function, amyloplasts are also known to play a role in the production of abscisic acid and carotenoids.108,109 In contrast, differentiation of proplastids, and not chloroplasts, into elaioplasts and proteinoplasts is non-reversible (Fig. 3). Elaioplasts plays a key role in storage and biosynthesis of lipids and oils including terpenoids, which in citrus fruits are then exported into secretory pockets, greatly affecting the aroma and taste.110,111 Furthermore, elaioplasts are also biosynthetic active plastids harboring the key upstream enzymes of the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway, which provides the precursors to synthesize mono- and diterpenes. Proteinoplasts (or proteoplasts) are defined as the site of protein and lipoprotein storage in the plant cell and these plastids are neither known to encompass biosynthesis of nor to act as storage sites for phytochemicals. The diversity of plastids and their specialized roles in different plant cell types throughout plant development emphasizes the dynamics and complexity of plant metabolism. In senescing leaves gerontoplasts are generated as the last ontogeny stage of chloroplasts (Fig. 3). These plastids no longer harbor functional DNA as resources are recycled and redistributed within the plant. How exactly the degradation of the thylakoid membrane systems occurs is not fully understood, but during degradation more and bigger plastoglobuli are formed.112–115
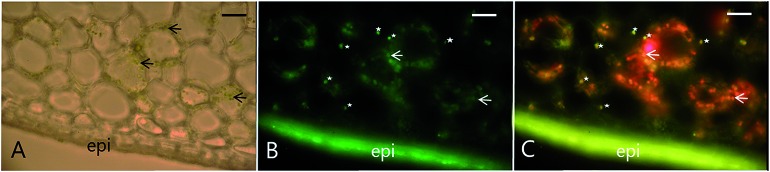
Recent studies on re-differentiated chloroplasts demonstrated that these may serve as biosynthetic units as well as storage site of phenylpropanoids. Based on studies in the vanilla (Vanilla planifolia) pod that focused on their ability to accumulate vanillin glucoside, a phenylpropanoid derived defense compound, these plastids were named phenyloplasts (Fig. 3). The transition from chloroplasts to phenyloplasts proceeds by a decline of photosynthetic capacity, thylakoid lamellae and chlorophyll content paralleled by accumulation of high concentrations of phenylpropanoid-derived glucosides.116,117 The molecular mechanisms that trigger the re-differentiation from chloroplast to phenyloplasts are currently unknown. The vanilla pod is the prime plant source of vanillin and the site of vanillin glucoside biosynthesis and storage.118–121 Recently, it was demonstrated that vanillin is biosynthesized in the chloroplasts and phenyloplasts by the enzyme VpVAN catalyzing the conversion of ferulic acid and its glucoside into vanillin and vanillin glucoside, respectively (Fig. 4).121 Toxic vanillin is stored as its non-toxic glucoside.122 Using multiple cell imaging approaches, it was discovered that vanillin glucoside was progressively accumulated in the inner volume of the phenyloplasts ultimately filling the entire phenyloplast at pod maturity.117 Vanillin β-glucosidase catalyzes hydrolysis of vanillin glucoside giving rise to formation of vanillin. This enzyme was reported to localize in the corona around re-differentiating chloroplasts, most likely in the lumen between the inner and outer chloroplast membranes.117 The spatial separation provides an efficient two-component based defense system against herbivores. The biosynthesis and storage of vanillin glucoside in phenyloplasts may represent a classical case of sub-cellular sequestration of phenolic compounds in specialized cellular compartments. Using Raman spectroscopy imaging of cross-sections of etiolated sorghum seedlings,123 the cyanogenic glucoside dhurrin was found to localize in the apoplast or cytoplasm, in the latter case most likely in phenyloplasts as previously suggested by Brillouet et al. (2014) based on indirect histochemical data and electron microscopy images.117,124 Storage of dhurrin in phenyloplasts may have developed as a consequence of the chemical arms-race leading to high intracellular concentration of phytochemicals,125 which accumulate up to 30% of the dry weight in the tip of etiolated sorghum seedlings corresponding to molar concentrations.49,124,126
Fig. 4. Colocalization of VpVAN, the first committed step in vanillin biosynthesis, in chloroplasts and phenyloplasts in 7-month-old Vanilla planifolia pods. Panel (A) shows light microscopy image of transverse section of the vanilla pod localizing chlorophyll containing green chloroplasts (black arrows indicating selected chloroplasts). Panel (B) shows immunolocalization of VpVAN (in green) in either chloroplasts or phenyloplasts. VpVAN was visualized by fluorescence microscopy using specific antibodies raised against VpVAN. Panel (C) shows the localization of VpVAN in chloroplasts (white arrows) and phenyloplasts (white stars) using fluorescence microscopy filter settings for simultaneous detection of both VpVAN (via FITC) and chloroplasts by chlorophyll auto-fluorescence. Chlorophyll (Chl) auto-fluorescence is shown in red indicating the localization of chloroplasts. Re-differentiating chloroplast are observed in yellow as the Chl auto-fluorescence is lower and phenyloplasts display no Chl auto-fluorescence and appear green. Scale bars equal to 100 μm. epi, epicarp. Figure adapted from Gallage et al. 2018, Plant & Cell Physiology.121.
4. Formation of micro-compartments by liquid–liquid phase separation
The ability of plants to biosynthesize and accumulate phytochemicals demonstrates an evolutionary advantage in the continuous arms race to fend off predators and attract pollinators. The bioactivity and toxicity of many phytochemicals make them targets for metabolic inactivation to prevent auto-toxicity. Their safe storage often involves spatial separation of phytochemicals and hydrolyzing enzymes.127 In addition to classical compartments, membrane-less microcompartments assembled through LLPS present a more dynamic approach for compartmentalization. Membrane-less compartments were initially discovered in mammalian systems as Cajal bodies,128 P granules129 and as the nucleolus in the nucleus.130 However, the phenomenon of cytosolic demixing of small molecules is becoming increasingly important in the understanding of how cells organize their metabolism. Essentially, multivalent interactions of biomolecules drive the LLPS and formation of cytosolic functional condensates or droplet compartments.131 Such droplets may be an additional approach for accumulation of high concentrations of phytochemicals. The dynamic structures of membrane-less compartments allow them to swiftly dissolve upon environmental stimuli or tissue damage and therefore provide increased functional plasticity as compared to more rigid membrane confined compartments.132
4.1. Micro-compartments composed of natural deep eutectic solvents (NADESs)?
Many phytochemicals are poorly soluble in the aqueous and lipid phases. NAtural Deep Eutectic Solvents (NADESs) have been proposed to constitute a third intracellular solvent phase.133 NADES are composed of natural compounds including main plant cell constituents such as sugars, amino acids, choline and organic acids. When these crystalline components are mixed in the right stoichiometric proportions, they form a viscous liquid. This applies for a mixture of glucose : fructose : sucrose in a 1 : 1 : 1 molar ratio as known from honey and for a mixture of glucose : tartaric acid in a 1 : 1 molar ratio as found in raisins. Despite an almost complete removal of water, raisins maintain a liquid phase due to formation of intramolecular hydrogen bonds. This results in a high melting point depression causing the solids to liquefy and, in many cases, to remain fluid at room temperature. NADES are excellent solvents for water insoluble phytochemicals such as anthocyanins and flavonoids133,134 and has therefore been used as green solvents for decades in the chemical industry for chemical synthesis and isolation of phytochemicals.
In respect to phytochemicals, the properties of NADES could serve to extend plant plasticity; (i) NADES has almost no vapor pressure and thus retains water during desiccation caused by drought stress and heat waves; (ii) NADES are eminent solvents for non-water-soluble phytochemicals and for macromolecules like proteins, RNA, DNA and cell wall polymers; (iii) formation of NADES could be driven by LLPS assisted by their high density and viscosity, with a tailored composition to effectively solubilize specific phytochemicals present in the cell type.133,135 A recent study also used NADES to solubilize and extract vanillin from cured vanilla pods.136 The curing process of vanilla pods disrupts cell integrity resulting in β-glucosidase catalyzed hydrolysis of the accumulated vanillin glucoside into the flavor molecule vanillin. The in vitro solubility of vanillin in NADES composed of lactic acid : 1,2-propanediol at a 1 : 1 molar ratio was 620 mg ml–1 resembling the solubility in methanol (633 mg ml–1). For comparison the solubility of vanillin in water is 30 mg ml–1.136 These observations support a function of NADES as a potential native solvent inside the phenyloplasts enabling accumulation of vanillin glucoside at 4 M (corresponding to ∼1800 mg ml–1) concentrations without precipitation or crystallization in fresh vanilla pods.117,121 Interestingly, the extractability of vanillin from cured vanilla pods using NADES was further improved by addition of 25–60% water. Upon addition of water above 50% the NADES properties are gradually abolished.135In vivo, differences in the water content in NADES droplets could control the activity of enzymes co-localized with their substrates in NADES137 and thus form an inert defense micro-environment ready to burst as a result of tissue damage and demixing.
4.2. Vacuolar microcompartments
The plant vacuole is a multifunctional compartment and a major site for sequestration of general metabolites, proteins, ions, and phytochemicals.138 Some of these biomolecules may be organized within sub-compartments assigned as inclusions. This applies to some phenylpropanoid derived anthocyanins that serve as flower pigments. Depending on both the plant species and tissue, different anthocyanins accumulate inside the vacuole, either as soluble compounds or as dense droplets with intense coloration termed anthocyanic vacuolar inclusions (AVIs).139–141 The inclusions vary in size and structure depending on the plant species, ranging from 0.5–15 μm in grapevine (Vitis vinifera L.),142 3–6.5 μm in arabidopsis143 and to app. 11–15 μm in transgenic N. benthamiana.144 Vacuolar localization provides protection from oxidation and increased solubility at low pH.140,141 Formation of AVIs may involve vesicular trafficking, vacuolar membrane transporter-mediated import and micro-autophagy of cytoplasmic anthocyanin condensates.139,143 However, factors controlling aggregation into the droplet-like structures are currently not known. In vitro experiments simulating AVI formation demonstrated that high salt concentrations and pH as well as aromatic acylation of the cyanidin-backbone are key factors involved.144 At high ionic strength and pH above 4.5, coumaroylation of the 5-OH-position instead of glycosylation was found to be especially important for intramolecular stacking and condensation.144In vivo, AVIs have been suggested to form inside vesicles in the cytosol followed by fusion with the vacuole similarly to the micro-autophagy model.144 A common denominator for the AVI formation models is that aromatic acylated anthocyanins are able to pack tightly together in aggregates due to intra- and inter-molecular associations145 and the high ionic strength of NADES33,146 facilitates self-stacking of anthocyanin molecules145 resulting in LLPS. Anthocyanins are easily extracted from plant tissues in NADES and display increased stability comparable to using acidified organic solvents.147 Similarly, in sorghum the phytoalexin 3-deoxyanthocyanidins have been shown to self-assemble through LLPS into dense cytosolic droplets upon fungal attack.148 These droplets, starting at 1 μm in size, fuse to form larger spherical inclusions of 20 μm that destabilizes the plasma membrane in the infected zone to combat infection.148 Further studies are needed to elucidate factors promoting this cytosolic demixing. Interestingly, the biosynthetic machinery of flavonoid 3-deoxyanthocyanidins, localized on the cytosolic side of the ER, is in close proximity to the initiation of condensation similar to mammalian LLPS composed of RNA and nuclear proteins in the nucleus.131,132 This adds to the importance of ER dynamics and interactions between cellular compartments12 and provides a synthetic biology tool-set for production of specific phytochemicals.
4.3. ER-derived microcompartments
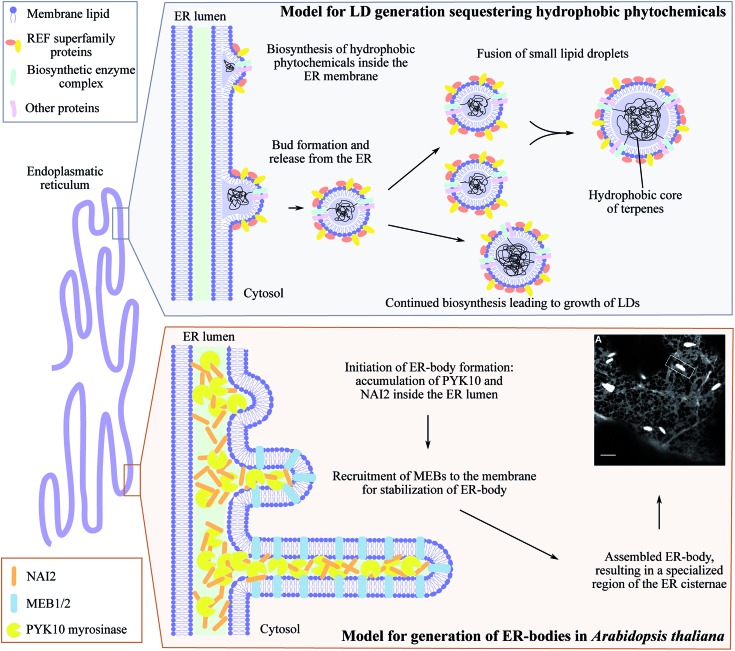
The ER is a highly dynamic compartment with many essential roles including the formation of microcompartments such as ER bodies (Fig. 5). These have diverse functions and appearances from small spherical inclusions with diameters of ∼0.2–1 μm to larger non-spherical structures with a size of 1 μm × 10 μm as found in Brassica species.149,150 In the brassica plant arabidopsis, ER bodies are constitutively found in seedlings and roots of adult plants and only stress-inducible in rosette leaves.151 Although several ER body mutants have been characterized, heterologous formation of ER bodies in non-ER body forming plants like N. benthamiana has not been achieved.150 In arabidopsis, the soluble NAI2 protein governs the specific accumulation of the most abundant β-glucosidase PYK10 myrosinase required for ER body formation (Fig. 5). nai2 mutants lack ER bodies in roots and seedlings resulting in uniform distribution of PYK10 in the ER.151 NAI2 facilitates the recruitment of two structural membrane proteins termed membrane of ER body, MEB1 and MEB2, that regulate the formation of ER bodies and inclusion of PYK10. Interestingly, only NAI2 and PYK10 are essential for ER body formation. The exact function of PYK10 loaded ER bodies remains to be elucidated. However, recently PYK10 was reported to hydrolyze indole glucosinolates and to release toxic mustard oil.150,152 Indole glucosinolates are biosynthesized at the ER cytosolic surface and functional networks between the indole glucosinolate-specific biosynthesis enzyme CYP83B1 and several vesicle proteins have been shown by protein–protein interaction studies.153 In addition, PYK10 showed strong co-expression with genes encoding both ER body proteins and indole glucosinolate biosynthetic enzymes.152 We speculate that both indole glucosinolates and their specific β-glucosidase PYK10 may be co-stored within ER bodies. Reversible inactivation of PYK10 may be achieved by controlling the pH inside ER bodies.152 This provides an excellent two-component defense system for release of mustard oil upon disruption of the ER body. In addition to glucosinolates in Brassica species, cyanogenic glucoside catabolism appears to be organized in a similar fashion. Cytosolic droplets or protein bodies (app. 1–3 μm) containing specific β-glucosidase activity have been observed in bitter almonds (Prunus dulcis),154,155 mature seeds of black cherry (Prunus serotina)156 and plums (Prunus domestica).157 Indeed, the combination of ER bodies and NADES pose an intriguing approach for storage of high concentrations of phytochemical defense compounds and their activating enzymes in an inert environment. However, it remains to be determined whether the cyanogenic glucosides are co-stored with their activating enzymes in these ER body like droplets.
Fig. 5. ER-derived microcompartments, lipid droplets (LDs) and ER-bodies. Top panel: shows the model for lipid droplets sequestering hydrophobic phytochemicals inspired by Laibach et al. 2015, Journal of Biotechnology.160 During biosynthesis, the hydrophobic phytochemical such as natural rubber accumulate inside the ER membrane leaflet causing bud formation and release of LD confined by an ER membrane monolayer. Small LDs may fuse to larger LDs at the same time as continued biosynthesis may occur resulting in growth of LDs. Lower panel: illustrates the development of ER-bodies in Arabidopsis thaliana in which the myrosinase PYK10 accumulates inside the ER lumen together with the soluble protein NAI2. This results in bud formation and triggers the recruitment of the membrane proteins MEB1 and MEB2 that interacts with NAI2, elongating the bud into the rod-shaped ER-body (1 μm× 10 μm) highly enriched with PYK10 and other β-glucosidases. Confocal image of ER-localized GFP (grey ER-network) in ER-bodies (white dilated ER cisternae) in Arabidopsis thaliana is adapted from Nakano et al. 2014, Frontiers in Plant Science.150.
Lipid droplets (LDs) are other ER-derived inclusions found throughout the plant kingdom including liverworts and higher plants.158,159 In higher plants, LDs can be divided into two types depending on their structural proteins: oleosin-based lipid droplets (OLDs) and non-oleosin-based lipid droplets (NOLDs). OLDs storing triglycerides and predominantly found in seeds serving as energy reserves during germination. The NOLDs are found in other tissues such as leaves and roots are 8–30 μm in size depending on the plant species and may serve as a storage site of lipophilic phytochemicals used for defense.159,160 Rubber particles are the best characterized NOLDs that contain natural rubber, high molecular weight polymers of cis-1,4-isoprene units, in laticifer cells of certain plants such as the rubber tree (Hevea brasiliensis), dandelion (Taraxacum Brevicorniculatum) and lettuce (Lactuca sativa). Rubber particles are formed by ER-anchored rubber synthesizing protein complexes depositing rubber polymers inside the ER leaflet (Fig. 5). Budding of the ER membrane leads to formation of rubber particle surrounded by an ER-derived lipid monolayer. The synthesis continues after particle formation resulting in different sizes of the rubber particles.160 Recent studies in dandelions identified Small Rubber Particle Proteins (SRPPs) belonging to the REF (rubber elongation factors) protein superfamily as important factors for initiation and stabilization of rubber particles.160–162 The SRPPs, like REFs interacts with the biosynthetic complex on the surface of the rubber particles. Heterologous expression in arabidopsis demonstrated that different SRPP isoforms displayed specific protein interactions, lipid preference, expression patterns and responses to abiotic stress providing heat and drought tolerance.161 Hence, lipid droplet associated proteins facilitate metabolic plasticity in response to environmental challenges. In non-rubber producing plants, SRPP-like proteins are known as Lipid Droplet Associated Proteins (LDAPs) involved in LD formation for storage of fatty acids in seeds.163,164 In arabidopsis, these LDs are turned into micro-factories of the phytoalexin 2-hydroxyoctadecatrienoic acid upon fungal attack.165,166
In contrast to higher plants, LDs in liverworts (size 1–30 μm) are reported to play a unique role as dynamic micro-compartments for both energy storage and sequestration of phytochemicals, mainly sesqui- and diterpenoids.158,167 Characterization of these terpenoids, many of which are unique to liverworts, will shed new light on the enormous diversity and specialization of phytochemicals during plant evolution. Recent, a comparison of putative di-, sesqui- and monoterpene synthases from the liverwort Marchantia polymorpha with those known from higher plants showed that liverwort genes encoding mono- and sesquiterpene synthases resembles those found in fungi more than those found in higher plants.168 These terpene synthases lack some of the conserved motifs present in those from higher plants. This may explain the unique terpenoid metabolite profile of liverworts and offers an experimental system to study the evolution and diversification of terpenoids and the plasticity of plant specialized metabolism in general. This includes the formation of LDs as micro-compartments for storage of terpenoids. Terpenoids from higher plants attract much attention based on their importance as drugs or drug leads.169 This includes the diterpenoid forskolin that is biosynthesized and stored in LDs (∼5–9 μm in size) in the root cortex of Coleus forskohlii.95,170 Little is known about the biogenesis of LDs and the mechanisms controlling their formation and stability. Characterization of key factors regulating their formation will provide an additional synthetic biology tool-set for heterologous production of lipophilic and volatile high value phytochemicals.
Other terpenoids may also be stored in the roots although without forming lipid droplets. This is observed in Tripterygium species producing the triterpenoid triptolide and the diterpenoid celastrol.171 Celastrol accumulates in high amounts in the periderm of the root and colors the root tissue orange. Using MALDI-imaging and microscopy, co-localization of celastrol and suberized cells was observed suggesting the cell wall as the site of storage due to its lipophilic nature.
Finally, extracellular glands pose an efficient approach for storage of phytochemicals, such as glandular trichomes encapsulating the terpenoid-containing essential oils found in many herbs.172 In the Eucalyptus genus, the leaves contain numerous embedded glands (50–120 μm in size) that are extracellular compartments for storage of large amounts of volatile mono- and sesquiterpenoids173,174 (Fig. 6). A barrier of oleuropeyl glucose esters confines the mono- and sesquiterpenoids inside the extracellular glands resulting in safe storage of potentially reactive oils.173,174
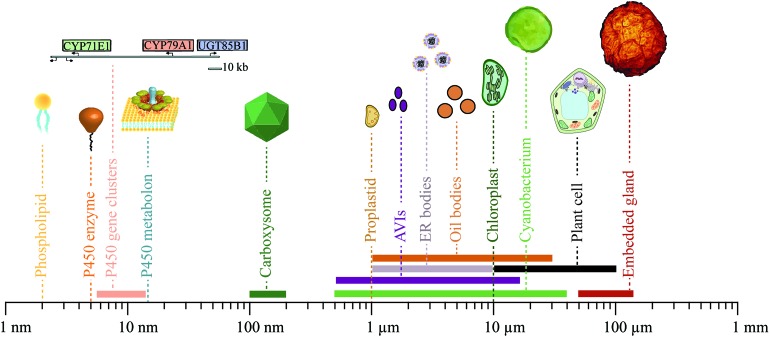
Fig. 6. Size distribution of components, modules and compartments involved in phytochemical metabolism and storage. Sizes range from 2 nm for a single phospholipid, the main component of biological membranes, to 120 μm for an embedded gland surrounded by multiple cells. The geometrical dimensions shown are in some cases somewhat arbitrary, e.g. because a gene sequence may be in a supercoiled configuration or in an open configuration as when transcribed.
5. Confined biosynthesis and storage as an eminent synthetic biology toolbox
Synthetic biology is a new paradigm that applies advanced engineering principles to the fundamental components of biology based on functional integration of Nature's building blocks in new combinations.175 Confinement of biosynthetic modules in enzyme complexes, or in subcellular compartments is an efficient way to orchestrate the metabolic grid in plants. However, the complexity of regulatory networks still poses a challenge for the successful exploitation of these strategies in planta. Within synthetic biology, specific modules may be transferred from one organism to another and thereby mimicking Natures' rational design to improve the production of specific compounds. The modularity of biological systems provides an endless toolbox for engineering optimized or entirely new biological systems.92,176–178 This involves programmable metabolons using synthetic scaffold proteins, engineering proteinaceous compartments and extended use of existing compartments. Here we highlight a few recent synthetic biology applications and perspectives in engineering cellular organization, dynamics and compartmentalization to produce and store phytochemicals.
5.1. The plant plasticity toolbox
The physiology and biochemistry of plants is highly complex and sophisticated at all levels and is constantly being optimized and shaped by evolution. Consequently, the biosynthesis, transport and storage of phytochemicals in a plant cell is not parsimonious in construction, containing an abundance of linear and non-linear feedback regulations and many different information processing modules to orchestrate dynamic responses and robust solutions to a sessile life style. To profit from their ability to biosynthesize a plethora of phytochemicals, the correct interplay at many different cellular levels and between many different modules is essential. Dissection of these dynamic and complex functional networks into the basic functions of the individual modules involved is a challenge. The continued advances in research within this area will provide access to an unprecedented library of synthetic biology modules ranging from small molecules to complex ordered aggregations of specific cell types varying in geometrical sizes from around a few nm to the 100 μm range (Fig. 6).
Biosynthesis of phytochemicals is initially organized at the genomic level. A gene cluster encoding an entire biosynthetic pathway in a fully packed chromosome, may only extend chromosome length by 5.5–10.5 nm.73,76,179 Single genes as well as clusters may thus be used as functional modules. The geometrical size of a P450 enzyme is roughly 5 nm, but when assembled in metabolons is estimated to be roughly 15 nm based on the combination of P450s and POR.6 Cyanobacteria produce proteinaceous compartments, such as the carboxysomes, which are icosahedral structures surrounded by a protein shell with geometrical sizes of 100–200 nm.180,181 The geometrical dimensions of two of the canonical membrane confined compartments in the plant cell, the proplastids and chloroplasts, are roughly 1 and 10 μm, respectively.82 The geometrical sizes of the more deformative inclusions are more variable e.g. 0.5–15 μm for anthocyanic vacuolar inclusions (AVIs),142–144 1–10 μm for ER bodies,149,150 1–30 μm for lipid droplets158,159,170 and 50–120 μm for tissue embedded glands encapsulated by several cells.173,174 Finally, photosynthetic cells as light-driven carbon dioxide utilizing production entities for production of phytochemicals such as cyanobacteria and plant cells vary in sizes from 0.5–40 μm182 and 10–100 μm, respectively.
5.2. Engineering modules for production and storage of phytochemicals
At the cellular level, optimal confinement of enzymes serves to increase their specificity, stimulate flux and prevent undesired metabolic crosstalk. Use of synthetic scaffold proteins offers the opportunity to mimic the formation of metabolons by programmable assembly of desired stoichiometric ratios of enzymes catalyzing consecutive steps in a biosynthetic pathway in to facilitate efficient substrate channeling.183,184 Scaffold proteins play a key role in signaling pathways for specific activation of downstream effectors, where enzymes are tethered to the scaffold through highly specific PDZ domains with nM affinity.185 This approach has been used to engineer the native yeast mitogen-activated protein kinase (MAPK) cascade by altering the binding sites of the Ste5 scaffold protein resulting in an adjustable response circuit186 which may find general use in improving substrate channeling for production of specialized metabolites. In 2009, the core enzymes of the yeast mevalonate pathway (AtoB, HMGS and HMGR) were tethered to a synthetic scaffold protein with three distinct docking sites using E. coli as the heterologous host. The option to control the stoichiometry between the individual enzymes tethered to the scaffold protein offered a 77-fold increase in mevalonate production compared to untethered proteins. A similar programmable platform was used for the production of resveratrol in yeast by coupling the 4-CL enzyme to a stilbene synthase reaching a 5-fold increase in production.187 Scaffold proteins may also be used to facilitate the production of new-to-nature compounds.
Biosynthesis of phytochemicals is highly complex, as described in this review, and therefore requires sophisticated approaches to achieve high yield production in heterologous hosts. Elucidation of key factors stimulating assembly of biosynthetic pathways for production of phytochemical pathways into metabolons may enable rational design of metabolons with enhanced stability and facilitate incorporation of other enzymes for channeled production of new metabolites. The newly discovered MSPB scaffold protein involved in phenylpropanoid biosynthesis60 provides one of the first characterized scaffold proteins involved in biosynthesis of phytochemicals and a valuable future tool for more efficient production of phenylpropanoid derived compounds. A key question remains on the role of the lipid bilayer composition on formation of metabolons. This may pose a key challenge when transferring entire multienzyme pathways containing membrane-bound members from one organism to another and possibly require engineering of the lipid composition to facilitate a perfect biosynthetic environment.
In some cases, the production of desired target compounds requires a more stringent intracellular organization where both enzymes and substrates are concentrated. In recent years, some research has focused on engineering minimal cells or protocells, synthetic compartments and vesicles to exploit these systems as micro-factories for production of chemicals.188 Metabolic engineering of intracellular compartments for biosynthesis and storage of phytochemicals may assist in preventing unwanted metabolic cross talk and competition for resources with native endogenous pathways.97,188,189 Metabolic compartmentalization may also serve to sequester and store otherwise toxic intermediates.5,190 This may be achieved by encapsulation of entire biosynthetic pathways in membrane-bound or proteinaceous compartments. Targeting of the biosynthetic enzymes for production of the sesquiterpenoid valencene from Citrus sinensis to the yeast mitochondria resulted in titers with an 8-fold increase in valencene production.191 Bacterial proteinaceous compartments provide a key synthetic biology tool for introducing synthetic compartments in heterologous hosts.192 This enables complete control of the cargo with limited inherent control from the host organism. Carboxysomes are present in cyanobacteria and function as a CO2 fixing compartment where both CO2 and ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) are concentrated. This is important because RuBisCO requires high concentrations of substrate for efficient turnover. Expression in E. coli of the ten genes encoding the constituents of the carboxysome, including various shell proteins, RuBisCO and carbonic anhydrase, resulted in formation of icosahedral structures that were able to fix CO2.193 This demonstrates the modularity of biological compartments, which is a central part of the synthetic biology vision.
Inspired by Nature and especially the dynamics of plant metabolism and strategies for biosynthesis and storage of complex phytochemicals including LLPS and formation of micro-compartments enables a more efficient production in heterologous hosts. For example, a 2-fold increase in ethyl acetate production in yeast was achieved by sequestering lipophilic phytochemicals into LD through the design of LD-anchored metabolons using synthetic protein scaffolds composed of plant oleosins and cohesin-dockerin proteins.194 In an alternative approach, chloroplast LDs were engineered to accumulate the high value triterpene squalene, the precursor of both more complex hydroxylated triterpenoids and plant sterols.195 The synthetic LDs were made by expression of the oleosin core structure fused with an N-terminal chloroplast targeting peptide and overexpression of the enzymes farnesyl diphosphate synthase and squalene synthase in Nicotiana tobacum, yielding 2.6 mg squalene per gram fresh weight with no influence on plant fitness.195 Such strategies could prove useful for production of terpenoids in plastids e.g. chromoplasts, which contain specialized LDs, the plastoglobules, or the lipid droplets inside elaioplasts to biosynthesize and accumulate desired high value terpenoids.97
5.3. A bright green future of synthetic biology
Production of phytochemicals in photosynthetic organisms including higher plants are challenging due to their complexity compared to unicellular production hosts e.g. Saccharomyces cerevisiae and E. coli. Realization of the full potential of photosynthetic organisms as heterologous hosts for effective, light driven carbon dioxide-based production of selected target molecules requires a detailed understanding of their complexity at the organismal level. The downstream challenges currently encountered with respect to purification of a compound of interest from a large complex mixture of different molecules with similar structures and physicochemical properties, may within the next three decades be turned into the advantage of having available photosynthetic cell based production systems producing large quantities of almost any desired compound. The plasticity of photosynthetic organisms as observed in Nature is then embedded in more specialized uses as environmental benign factories and storehouses of valuable phytochemicals, the formation of which is catalyzed by channeled pathways to avoid undesired metabolic cross-talk. The products formed will be targeted to specific cell types or storage compartments to make isolation easy.97,192,196 In parallel with the development of photosynthesis based production systems discussed above, research efforts will be directed towards further improvements of the less complex organisms such as fungal and bacterial hosts currently used for production of phytochemicals at industrial scale.177,197,198 In the long term, the inherent ability of photosynthetic organisms to produce a plethora of phytochemicals may be a decisive advantage with respect to establishing robust, efficient and environmentally benign production systems. In this context, advances in the development of technological platforms for characterization of subcellular structures with sub-nanometer resolution offer new opportunities for the specific production of known and new-to-nature phytochemicals in confined compartments using targeted synthetic biology approaches.
6. Conflicts of interest
The authors declare no competing financial interest.
Acknowledgments
The authors acknowledge financial support from the VILLUM Research Center for Plant Plasticity; by the bioSYNergy program of Center for Synthetic Biology (University of Copenhagen Excellence Program for Interdisciplinary Research); by a European Research Council Advanced Grant to B. L. M. (ERC-2012-ADG_20120314). T. L. is recipient of a fellowship awarded by the Novo Nordisk Foundation (project no. NNF17OC0024886). C. C. H. was supported by a PhD fellowship provided through Young Investigator Program fellowship from the VILLUM Foundation granted to E. H. J. Neilson (grant number 13167).
References
- Moghe G. D., Last R. L. Plant Physiol. 2015;169:1512–1523. doi: 10.1104/pp.15.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasling J. D., Mendoza A., Baran P. S. Nature. 2012;492:188–189. doi: 10.1038/492188a. [DOI] [PubMed] [Google Scholar]
- Møller B. L., Conn E. E. J. Biol. Chem. 1980;255:3049–3056. [PubMed] [Google Scholar]
- Winkel B. S. Annu. Rev. Plant Biol. 2004;55:85–107. doi: 10.1146/annurev.arplant.55.031903.141714. [DOI] [PubMed] [Google Scholar]
- Jørgensen K., Rasmussen A. V., Morant M., Nielsen A. H., Bjarnholt N., Zagrobelny M., Bak S., Møller B. L. Curr. Opin. Plant Biol. 2005;8:280–291. doi: 10.1016/j.pbi.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Laursen T., Borch J., Knudsen C., Bavishi K., Torta F., Martens H. J., Silvestro D., Hatzakis N. S., Wenk M. R., Dafforn T. R., Olsen C. E., Motawia M. S., Hamberger B., Møller B. L., Bassard J. E. Science. 2016;354:890–893. doi: 10.1126/science.aag2347. [DOI] [PubMed] [Google Scholar]
- Ellis R. J. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- Rivas G., Minton A. P. Trends Biochem. Sci. 2016;41:970–981. doi: 10.1016/j.tibs.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C. Science. 1978;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- Hrazdina G., Jensen R. A. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992;43:241–267. [Google Scholar]
- Sweetlove L. J., Fernie A. R. Annu. Rev. Plant Biol. 2013;64:723–746. doi: 10.1146/annurev-arplant-050312-120233. [DOI] [PubMed] [Google Scholar]
- Phillips M. J., Voeltz G. K. Nat. Rev. Mol. Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler A., Yoshimoto M., Luginbuhl S., Mavelli F., Walde P. Nat. Nanotechnol. 2016;11:409–420. doi: 10.1038/nnano.2016.54. [DOI] [PubMed] [Google Scholar]
- Aguilera-Gomez A., Rabouille C. Dev. Biol. 2017;428:310–317. doi: 10.1016/j.ydbio.2017.03.029. [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart D., Feyereisen R. Genome Biol. 2000;1:reviews3003. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shephard E. A., Phillips I. R., Bayney R. M., Pike S. F., Rabin B. R. Biochem. J. 1983;211:333–340. doi: 10.1042/bj2110333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozalp C., Szczesna-Skorupa E., Kemper B. Drug Metab. Dispos. 2005;33:1382–1390. doi: 10.1124/dmd.105.005538. [DOI] [PubMed] [Google Scholar]
- Backes W. L., Kelley R. W. Pharmacol. Ther. 2003;98:221–233. doi: 10.1016/s0163-7258(03)00031-7. [DOI] [PubMed] [Google Scholar]
- Peterson J. A., Ebel R. E., O'Keeffe D. H., Matsubara T., Estabrook R. W. J. Biol. Chem. 1976;251:4010–4016. [PubMed] [Google Scholar]
- Wädsater M., Laursen T., Singha A., Hatzakis N. S., Stamou D., Barker R., Mortensen K., Feidenhans'l R., Møller B. L., Cardenas M. J. Biol. Chem. 2012;287:34596–34603. doi: 10.1074/jbc.M112.400085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen T., Jensen K., Møller B. L. Biochim. Biophys. Acta, Proteins Proteomics. 2011;1814:132–138. doi: 10.1016/j.bbapap.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Bavishi K., Li D., Eiersholt S., Hooley E., Petersen T., Møller B. L., Hatzakis N., Laursen T. Sci. Rep. 2018;8:6817. doi: 10.1038/s41598-018-24922-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere P. A. Trends Biochem. Sci. 1985;10:109–110. [Google Scholar]
- Castellana M., Wilson M. Z., Xu Y., Joshi P., Cristea I. M., Rabinowitz J. D., Gitai Z., Wingreen N. S. Nat. Biotechnol. 2014;32:1011–1018. doi: 10.1038/nbt.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastmalchi M., Bernards M. A., Dhaubhadel S. Plant J. 2016;85:689–706. doi: 10.1111/tpj.13137. [DOI] [PubMed] [Google Scholar]
- Fujino N., Tenma N., Waki T., Ito K., Komatsuzaki Y., Sugiyama K., Yamazaki T., Yoshida S., Hatayama M., Yamashita S., Tanaka Y., Motohashi R., Denessiouk K., Takahashi S., Nakayama T. Plant J. 2018;94:372–392. doi: 10.1111/tpj.13864. [DOI] [PubMed] [Google Scholar]
- Lallemand B., Erhardt M., Heitz T., Legrand M. Plant Physiol. 2013;162:616–625. doi: 10.1104/pp.112.213124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassard J. E., Mutterer J., Duval F., Werck-Reichhart D. FEBS J. 2012;279:1576–1583. doi: 10.1111/j.1742-4658.2011.08312.x. [DOI] [PubMed] [Google Scholar]
- Bassard J. E., Richert L., Geerinck J., Renault H., Duval F., Ullmann P., Schmitt M., Meyer E., Mutterer J., Boerjan W., De Jaeger G., Mely Y., Goossens A., Werck-Reichhart D. Plant Cell. 2012;24:4465–4482. doi: 10.1105/tpc.112.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M., Jørgensen K., Schaller H., Pinot F., Møller B. L., Werck-Reichhart D., Bak S. Plant Cell. 2007;19:1473–1487. doi: 10.1105/tpc.106.045948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford H. Annu. Rev. Plant Biol. 1974;25:459–486. [Google Scholar]
- Achnine L., Blancaflor E. B., Rasmussen S., Dixon R. A. Plant Cell. 2004;16:3098–3109. doi: 10.1105/tpc.104.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen T., Møller B. L., Bassard J. E. Trends Plant Sci. 2015;20:20–32. doi: 10.1016/j.tplants.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Ralston L., Yu O. Phytochem. Rev. 2006;5:459–472. [Google Scholar]
- Bassard J. E., Møller B. L., Laursen T. Current Molecular Biology Reports. 2017;3:37–51. doi: 10.1007/s40610-017-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waki T., Yoo D., Fujino N., Mameda R., Denessiouk K., Yamashita S., Motohashi R., Akashi T., Aoki T., Ayabe S., Takahashi S., Nakayama T. Biochem. Biophys. Res. Commun. 2016;469:546–551. doi: 10.1016/j.bbrc.2015.12.038. [DOI] [PubMed] [Google Scholar]
- Andersen T. B., Hansen N. B., Laursen T., Weitzel C., Simonsen H. T. Mol. Phylogenet. Evol. 2016;98:21–28. doi: 10.1016/j.ympev.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Bertram N., Laursen T., Barker R., Bavishi K., Møller B. L., Cardenas M. Langmuir. 2015;31:8386–8391. doi: 10.1021/acs.langmuir.5b00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K., Møller B. L. Phytochemistry. 2010;71:132–141. doi: 10.1016/j.phytochem.2009.10.017. [DOI] [PubMed] [Google Scholar]
- Laursen T., Singha A., Rantzau N., Tutkus M., Borch J., Hedegard P., Stamou D., Møller B. L., Hatzakis N. S. ACS Chem. Biol. 2014;9:630–634. doi: 10.1021/cb400708v. [DOI] [PubMed] [Google Scholar]
- Bak S., Kahn R. A., Nielsen H. L., Møller B. L., Halkier B. A. Plant Mol. Biol. 1998;36:393–405. doi: 10.1023/a:1005915507497. [DOI] [PubMed] [Google Scholar]
- Jones P. R., Møller B. L., Høj P. B. J. Biol. Chem. 1999;274:35483–35491. doi: 10.1074/jbc.274.50.35483. [DOI] [PubMed] [Google Scholar]
- Kahn R. A., Bak S., Svendsen I., Halkier B. A., Møller B. L. Plant Physiol. 1997;115:1661–1670. doi: 10.1104/pp.115.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn R. A., Fahrendorf T., Halkier B. A., Møller B. L. Arch. Biochem. Biophys. 1999;363:9–18. doi: 10.1006/abbi.1998.1068. [DOI] [PubMed] [Google Scholar]
- Koch B. M., Sibbesen O., Halkier B. A., Svendsen I., Møller B. L. Arch. Biochem. Biophys. 1995;323:177–186. doi: 10.1006/abbi.1995.0024. [DOI] [PubMed] [Google Scholar]
- Kristensen C., Morant M., Olsen C. E., Ekstrøm C. T., Galbraith D. W., Møller B. L., Bak S. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1779–1784. doi: 10.1073/pnas.0409233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K. A., Tattersall D. B., Jones P. R., Møller B. L. Phytochemistry. 2008;69:88–98. doi: 10.1016/j.phytochem.2007.06.033. [DOI] [PubMed] [Google Scholar]
- Sibbesen O., Koch B., Halkier B. A., Møller B. L. J. Biol. Chem. 1995;270:3506–3511. doi: 10.1074/jbc.270.8.3506. [DOI] [PubMed] [Google Scholar]
- Halkier B. A., Møller B. L. Plant Physiol. 1989;90:1552–1559. doi: 10.1104/pp.90.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar N., Li L., Lu S., Khin N. C., Pogson B. J. Mol. Plant. 2015;8:68–82. doi: 10.1016/j.molp.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Bassard J. E., Halkier B. A. Phytochem. Rev. 2018;17:211–227. doi: 10.1007/s11101-017-9509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. C., Li Q., Shuford C. M., Liu J., Muddiman D. C., Sederoff R. R., Chiang V. L. Proc. Natl. Acad. Sci. U. S. A. 2011;108:21253–21258. doi: 10.1073/pnas.1116416109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan R. F., Jaradat T. T., Wurtzel E. T. Arch. Biochem. Biophys. 2007;458:146–157. doi: 10.1016/j.abb.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E. Nat. Methods. 2016;13:24–27. doi: 10.1038/nmeth.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Benlekbir S., Venkatakrishnan P., Wang Y., Hong S., Hosler J., Tajkhorshid E., Rubinstein J. L., Gennis R. B. Nature. 2018;557:123–126. doi: 10.1038/s41586-018-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S., Jamshad M., Knowles T. J., Morrison K. A., Downing R., Cant N., Collins R., Koenderink J. B., Ford R. C., Overduin M., Kerr I. D., Dafforn T. R., Rothnie A. J. Biochem. J. 2014;461:269–278. doi: 10.1042/BJ20131477. [DOI] [PubMed] [Google Scholar]
- Parmar M., Rawson S., Scarff C. A., Goldman A., Dafforn T. R., Muench S. P., Postis V. L. G. Biochim. Biophys. Acta, Biomembr. 2018;1860:378–383. doi: 10.1016/j.bbamem.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault J., Donlan J. A., Liko I., Hopper J. T., Gupta K., Housden N. G., Struwe W. B., Marty M. T., Mize T., Bechara C., Zhu Y., Wu B., Kleanthous C., Belov M., Damoc E., Makarov A., Robinson C. V. Nat. Methods. 2016;13:333–336. doi: 10.1038/nmeth.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganowsky A., Reading E., Allison T. M., Ulmschneider M. B., Degiacomi M. T., Baldwin A. J., Robinson C. V. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M., Ran X., Martin D. W., Liu C. J. Nat. Plants. 2018;4:299–310. doi: 10.1038/s41477-018-0142-9. [DOI] [PubMed] [Google Scholar]
- Chandler D. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- Simons K., Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Brignac-Huber L. M., Reed J. R., Eyer M. K., Backes W. L. Drug Metab. Dispos. 2013;41:1896–1905. doi: 10.1124/dmd.113.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordoba E., Salmi M., Leon P. J. Exp. Bot. 2009;60:2933–2943. doi: 10.1093/jxb/erp190. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H. K., Schwender J., Disch A., Rohmer M. FEBS Lett. 1997;400:271–274. doi: 10.1016/s0014-5793(96)01404-4. [DOI] [PubMed] [Google Scholar]
- Pateraki I., Heskes A. M., Hamberger B. Adv. Biochem. Eng./Biotechnol. 2015;148:107–139. doi: 10.1007/10_2014_301. [DOI] [PubMed] [Google Scholar]
- Perez-Sancho J., Tilsner J., Samuels A. L., Botella M. A., Bayer E. M., Rosado A. Trends Cell Biol. 2016;26:705–717. doi: 10.1016/j.tcb.2016.05.007. [DOI] [PubMed] [Google Scholar]
- Mehrshahi P., Johnny C., DellaPenna D. Trends Plant Sci. 2014;19:501–507. doi: 10.1016/j.tplants.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Møller B. L. Science. 2010;330:1328–1329. doi: 10.1126/science.1194971. [DOI] [PubMed] [Google Scholar]
- Kutchan T. M. Curr. Opin. Plant Biol. 2005;8:292–300. doi: 10.1016/j.pbi.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Nützmann H. W., Huang A., Osbourn A. New Phytol. 2016;211:771–789. doi: 10.1111/nph.13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M., Heinig U., Tzfadia O., Bhide A. J., Shinde B., Cardenas P. D., Bocobza S. E., Unger T., Malitsky S., Finkers R., Tikunov Y., Bovy A., Chikate Y., Singh P., Rogachev I., Beekwilder J., Giri A. P., Aharoni A. Science. 2013;341:175–179. doi: 10.1126/science.1240230. [DOI] [PubMed] [Google Scholar]
- Takos A. M., Knudsen C., Lai D., Kannangara R., Mikkelsen L., Motawia M. S., Olsen C. E., Sato S., Tabata S., Jørgensen K., Møller B. L., Rook F. Plant J. 2011;68:273–286. doi: 10.1111/j.1365-313X.2011.04685.x. [DOI] [PubMed] [Google Scholar]
- Winzer T., Gazda V., He Z., Kaminski F., Kern M., Larson T. R., Li Y., Meade F., Teodor R., Vaistij F. E., Walker C., Bowser T. A., Graham I. A. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- Frey M., Chomet P., Glawischnig E., Stettner C., Grun S., Winklmair A., Eisenreich W., Bacher A., Meeley R. B., Briggs S. P., Simcox K., Gierl A. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- Osbourn A. Plant Physiol. 2010;154:531–535. doi: 10.1104/pp.110.161315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hagel J. M., Chang L., Tucker J. E., Shiigi S. A., Yelpaala Y., Chen H. Y., Estrada R., Colbeck J., Enquist-Newman M., Ibanez A. B., Cottarel G., Vidanes G. M., Facchini P. J. Nat. Chem. Biol. 2018;14:738–743. doi: 10.1038/s41589-018-0059-7. [DOI] [PubMed] [Google Scholar]
- Bonk M., Hoffmann B., Von Lintig J., Schledz M., Al-Babili S., Hobeika E., Kleinig H., Beyer P. Eur. J. Biochem. 1997;247:942–950. doi: 10.1111/j.1432-1033.1997.00942.x. [DOI] [PubMed] [Google Scholar]
- Soll J., Alefsen H. Physiol. Plant. 1993;87:433–440. [Google Scholar]
- Mackenzie S., McIntosh L. Plant Cell. 1999;11:571–586. doi: 10.1105/tpc.11.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzlander M., Finkemeier I. Antioxid. Redox Signaling. 2013;18:2122–2144. doi: 10.1089/ars.2012.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K. Plant Cell. 1998;10:1971–1972. doi: 10.1105/tpc.10.12.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. Mol. Plant. 2009;2:1154–1180. doi: 10.1093/mp/ssp088. [DOI] [PubMed] [Google Scholar]
- Bruce B. D. Trends Cell Biol. 2000;10:440–447. doi: 10.1016/s0962-8924(00)01833-x. [DOI] [PubMed] [Google Scholar]
- Nielsen V. S., Mant A., Knoetzel J., Møller B. L., Robinson C. J. Biol. Chem. 1994;269:3762–3766. [PubMed] [Google Scholar]
- Kirk P. R., Leech R. M. Plant Physiol. 1972;50:228–234. doi: 10.1104/pp.50.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold F., vom Dorp K., Abraham M., Holzl G., Wewer V., Yilmaz J. L., Lager I., Montandon C., Besagni C., Kessler F., Stymne S., Dormann P. Plant Cell. 2012;24:2001–2014. doi: 10.1105/tpc.112.095588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Benning C. Biochem. Soc. Trans. 2012;40:457–463. doi: 10.1042/BST20110752. [DOI] [PubMed] [Google Scholar]
- Metraux J. P. Trends Plant Sci. 2002;7:332–334. doi: 10.1016/s1360-1385(02)02313-0. [DOI] [PubMed] [Google Scholar]
- DellaPenna D., Pogson B. J. Annu. Rev. Plant Biol. 2006;57:711–738. doi: 10.1146/annurev.arplant.56.032604.144301. [DOI] [PubMed] [Google Scholar]
- Von Wettstein D., Gough S., Kannangara C. G. Plant Cell. 1995;7:1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen-Ranberg J., Kongstad K. T., Nielsen M. T., Jensen N. B., Pateraki I., Bach S. S., Hamberger B., Zerbe P., Staerk D., Bohlmann J., Møller B. L., Hamberger B. Angew. Chem., Int. Ed. 2016;55:2142–2146. doi: 10.1002/anie.201510650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heskes A. M., Sundram T. C. M., Boughton B. A., Jensen N. B., Hansen N. L., Crocoll C., Cozzi F., Rasmussen S., Hamberger B., Hamberger B., Staerk D., Møller B. L., Pateraki I. Plant J. 2018;93:943–958. doi: 10.1111/tpj.13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munne-Bosch S., Alegre L. Plant Physiol. 2001;125:1094–1102. doi: 10.1104/pp.125.2.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateraki I., Andersen-Ranberg J., Jensen N. B., Wubshet S. G., Heskes A. M., Forman V., Hallstrom B., Hamberger B., Motawia M. S., Olsen C. E., Staerk D., Hansen J., Møller B. L., Hamberger B. eLife. 2017;6:e23001. doi: 10.7554/eLife.23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole L. W. Front. Cell. Dev. Biol. 2016;4:85. doi: 10.3389/fcell.2016.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor S. B., Behrendorff J., Nielsen A. Z., Jensen P. E., Pribil M. Essays Biochem. 2018;62:41–50. doi: 10.1042/EBC20170047. [DOI] [PubMed] [Google Scholar]
- Jarvis P., Lopez-Juez E. Nat. Rev. Mol. Cell Biol. 2013;14:787–802. doi: 10.1038/nrm3702. [DOI] [PubMed] [Google Scholar]
- Pogson B. J., Ganguly D., Albrecht-Borth V. Biochim. Biophys. Acta, Bioenerg. 2015;1847:1017–1024. doi: 10.1016/j.bbabio.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Villalon A., Gas E., Rodriguez-Concepcion M. Plant J. 2009;60:424–435. doi: 10.1111/j.1365-313X.2009.03966.x. [DOI] [PubMed] [Google Scholar]
- Sun T., Yuan H., Cao H., Yazdani M., Tadmor Y., Li L. Mol. Plant. 2018;11:58–74. doi: 10.1016/j.molp.2017.09.010. [DOI] [PubMed] [Google Scholar]
- Klee H. J., Giovannoni J. J. Annu. Rev. Genet. 2011;45:41–59. doi: 10.1146/annurev-genet-110410-132507. [DOI] [PubMed] [Google Scholar]
- Llorente B., D'Andrea L., Rodriguez-Concepcion M. Front. Plant Sci. 2016;7:263. doi: 10.3389/fpls.2016.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B., Martinez-Garcia J. F., Stange C., Rodriguez-Concepcion M. Curr. Opin. Plant Biol. 2017;37:49–55. doi: 10.1016/j.pbi.2017.03.011. [DOI] [PubMed] [Google Scholar]
- Camara B., Bardat F., Moneger R., Hebd C. R. Front. Plant Sci. 1982;294:649–652. [Google Scholar]
- Egea I., Barsan C., Bian W., Purgatto E., Latche A., Chervin C., Bouzayen M., Pech J. C. Plant Cell Physiol. 2010;51:1601–1611. doi: 10.1093/pcp/pcq136. [DOI] [PubMed] [Google Scholar]
- Roberts E. A. J. Appl. Phys. 1946;17:68. [Google Scholar]
- Paine J. A., Shipton C. A., Chaggar S., Howells R. M., Kennedy M. J., Vernon G., Wright S. Y., Hinchliffe E., Adams J. L., Silverstone A. L., Drake R. Nat. Biotechnol. 2005;23:482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- Zhai S., Xia X., He Z. Front. Plant Sci. 2016;7:1197. doi: 10.3389/fpls.2016.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizes M., Pauly G., Carde J. P., Marpeau A., Bernard-Dagan C. Planta. 1983;159:373–381. doi: 10.1007/BF00393177. [DOI] [PubMed] [Google Scholar]
- Yamasaki Y., Akimitsu K. J. Plant Physiol. 2007;164:1436–1448. doi: 10.1016/j.jplph.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Krupinska K., in The Structure and Function of Plastids, eds. R. R. Wise and J. K. Hoober, Springer, Dordrecht, 2007, vol. Advances in Photosynthesis and Respiration, pp. 433–449. [Google Scholar]
- Kutik J. Photosynthetica. 1998;35:481–505. [Google Scholar]
- Sitte P., in Biologie in unserer Zeit, Wiley Subscription Services, Inc., A Wiley Company, 1977, vol. 7, pp. 65–74. [Google Scholar]
- Zhu M., Lin J. J., Ye J. L., Wang R., Yang C., Gong J. L., Liu Y., Deng C. L., Liu P., Chen C. W., Cheng Y. J., Deng X. X., Zeng Y. L. Hortic. Res. 2018;5:6. doi: 10.1038/s41438-017-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillouet J. M., Romieu C., Schoefs B., Solymosi K., Cheynier V., Fulcrand H., Verdeil J. L., Conejero G. Ann. Bot. 2013;112:1003–1014. doi: 10.1093/aob/mct168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillouet J. M., Verdeil J. L., Odoux E., Lartaud M., Grisoni M., Conejero G. J. Exp. Bot. 2014;65:2427–2435. doi: 10.1093/jxb/eru126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odoux E., Brillouet J. M. Fruits. 2009;64:221–241. [Google Scholar]
- Odoux E., Escoute J., Verdiel J. L., Brillouet J. M. Ann. Bot. 2003;92:437–444. doi: 10.1093/aob/mcg150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallage N. J., Hansen E. H., Kannangara R., Olsen C. E., Motawia M. S., Jørgensen K., Holme I., Hebelstrup K. H., Grisoni M., Møller B. L. Nat. Commun. 2014;5:4037. doi: 10.1038/ncomms5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallage N. J., Jørgensen K., Janfelt C., Nielsen A. J. Z., Naake T., Dunski E., Dalsten L., Grisoni M., Møller B. L. Plant Cell Physiol. 2018;59:304–318. doi: 10.1093/pcp/pcx185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonchird C., Flegel T. W. Can. J. Microbiol. 1982;28:1235–1241. doi: 10.1139/m82-184. [DOI] [PubMed] [Google Scholar]
- Heraud P., Cowan M. F., Marzec K. M., Møller B. L., Blomstedt C. K., Gleadow R. Sci. Rep. 2018;8:2691. doi: 10.1038/s41598-018-20928-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders J. A., Conn E. E., Lin C. H., Stocking C. R. Plant Physiol. 1977;59:647–652. doi: 10.1104/pp.59.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon C. M., Langlois-Meurinne M., Saindrenan P. Trends Plant Sci. 2005;10:542–549. doi: 10.1016/j.tplants.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Kojima M., Poulton J. E., Thayer S. S., Conn E. E. Plant Physiol. 1979;63:1022–1028. doi: 10.1104/pp.63.6.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant A. V., Jørgensen K., Jørgensen C., Paquette S. M., Sanchez-Perez R., Møller B. L., Bak S. Phytochemistry. 2008;69:1795–1813. doi: 10.1016/j.phytochem.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Gall J. G. Nat. Rev. Mol. Cell Biol. 2003;4:975–980. doi: 10.1038/nrm1262. [DOI] [PubMed] [Google Scholar]
- Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A. A. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Pederson T. Cold Spring Harbor Perspect. Biol. 2011;3:a000638. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S. F., Lee H. O., Hyman A. A., Rosen M. K. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchaine E. M., Lu A., Neugebauer K. M. EMBO J. 2016;35:1603–1612. doi: 10.15252/embj.201593517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. H., van Spronsen J., Dai Y. T., Verberne M., Hollmann F., Arends I. W. C. E., Witkamp G. J., Verpoorte R. Plant Physiol. 2011;156:1701–1705. doi: 10.1104/pp.111.178426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y. T., van Spronsen J., Witkamp G. J., Verpoorte R., Choi Y. H. Anal. Chim. Acta. 2013;766:61–68. doi: 10.1016/j.aca.2012.12.019. [DOI] [PubMed] [Google Scholar]
- Dai Y. T., Witkamp G. J., Verpoorte R., Choi Y. H. Food Chem. 2015;187:14–19. doi: 10.1016/j.foodchem.2015.03.123. [DOI] [PubMed] [Google Scholar]
- González C. G., Mustafa N. R., Wilson E. G., Verpoorte R., Choi Y. H. Flavour Fragrance J. 2018;33:91–96. [Google Scholar]
- Aumiller, Jr. W. M., Keating C. D. Adv. Colloid Interface Sci. 2017;239:75–87. doi: 10.1016/j.cis.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Wink M. J. Exp. Bot. 1993;44:231–246. [Google Scholar]
- Tohge T., de Souza L. P., Fernie A. R. J. Exp. Bot. 2017;68:4013–4028. doi: 10.1093/jxb/erx177. [DOI] [PubMed] [Google Scholar]
- Pourcel L., Irani N. G., Lu Y. H., Riedl K., Schwartz S., Grotewold E. Mol. Plant. 2010;3:78–90. doi: 10.1093/mp/ssp071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham K. R., Gould K. S., Winefield C. S., Mitchell K. A., Bloor S. J., Boase M. R. Phytochemistry. 2000;55:327–336. doi: 10.1016/s0031-9422(00)00246-6. [DOI] [PubMed] [Google Scholar]
- Conn S., Franco C., Zhang W. Planta. 2010;231:1343–1360. doi: 10.1007/s00425-010-1139-4. [DOI] [PubMed] [Google Scholar]
- Chanoca A., Kovinich N., Burkel B., Stecha S., Bohorquez-Restrepo A., Ueda T., Eliceiri K. W., Grotewold E., Otegui M. S. Plant Cell. 2015;27:2545–2559. doi: 10.1105/tpc.15.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallam K., Appelhagen I., Luo J., Albert N., Zhang H., Deroles S., Hill L., Findlay K., Andersen O. M., Davies K., Martin C. Curr. Biol. 2017;27:945–957. doi: 10.1016/j.cub.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A., Brás N. F., Mateus N., Freitas V. d. New J. Chem. 2015;39:2602–2611. [Google Scholar]
- Dai Y., van Spronsen J., Witkamp G. J., Verpoorte R., Choi Y. H. J. Nat. Prod. 2013;76:2162–2173. doi: 10.1021/np400051w. [DOI] [PubMed] [Google Scholar]
- Dai Y., Rozema E., Verpoorte R., Choi Y. H. J. Chromatogr. A. 2016;1434:50–56. doi: 10.1016/j.chroma.2016.01.037. [DOI] [PubMed] [Google Scholar]
- Nielsen K. A., Gotfredsen C. H., Buch-Pedersen M. J., Ammitzboll H., Mattsson O., Duus J. O., Nicholson R. L. Physiol. Mol. Plant Pathol. 2004;65:187–196. [Google Scholar]
- Chrispeels M. J., Herman E. M. Plant Physiol. 2000;123:1227–1233. doi: 10.1104/pp.123.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano R. T., Yamada K., Bednarek P., Nishimura M., Hara-Nishimura I. Front. Plant Sci. 2014;5:73. doi: 10.3389/fpls.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Nagano A. J., Nishina M., Hara-Nishimura I., Nishimura M. Plant Cell. 2008;20:2529–2540. doi: 10.1105/tpc.108.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano R. T., Pislewska-Bednarek M., Yamada K., Edger P. P., Miyahara M., Kondo M., Bottcher C., Mori M., Nishimura M., Schulze-Lefert P., Hara-Nishimura I., Bednarek P. Plant J. 2017;89:204–220. doi: 10.1111/tpj.13377. [DOI] [PubMed] [Google Scholar]
- Nintemann S. J., Vik D., Svozil J., Bak M., Baerenfaller K., Burow M., Halkier B. A. Front. Plant Sci. 2017;8:2028. doi: 10.3389/fpls.2017.02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez R., Belmonte F. S., Borch J., Dicenta F., Møller B. L., Jørgensen K. Plant Physiol. 2012;158:1916–1932. doi: 10.1104/pp.111.192021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Perez R., Jørgensen K., Motawia M. S., Dicenta F., Møller B. L. Plant J. 2009;60:894–906. doi: 10.1111/j.1365-313X.2009.03997.x. [DOI] [PubMed] [Google Scholar]
- Swain E., Li C. P., Poulton J. E. Plant Physiol. 1992;100:291–300. doi: 10.1104/pp.100.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J. E., Li C. P. Plant Physiol. 1994;104:29–35. doi: 10.1104/pp.104.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. L., Sun Y., Zhu R. L. Crit. Rev. Plant Sci. 2013;32:293–302. [Google Scholar]
- Chapman K. D., Dyer J. M., Mullen R. T. J. Lipid Res. 2012;53:215–226. doi: 10.1194/jlr.R021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laibach N., Post J., Twyman R. M., Gronoyera C. S., Prufer D. J. Biotechnol. 2015;201:15–27. doi: 10.1016/j.jbiotec.2014.08.020. [DOI] [PubMed] [Google Scholar]
- Laibach N., Schmidl S., Muller B., Bergmann M., Prufer D., Gronover C. S. Plant J. 2018;93:1045–1061. doi: 10.1111/tpj.13829. [DOI] [PubMed] [Google Scholar]
- Laibach N., Hillebrand A., Twyman R. M., Prufer D., Gronover C. S. Plant J. 2015;82:609–620. doi: 10.1111/tpj.12836. [DOI] [PubMed] [Google Scholar]
- Huang M. D., Huang A. H. Plant Physiol. 2015;169:453–470. doi: 10.1104/pp.15.00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn P. J., James C. N., Gidda S. K., Kilaru A., Dyer J. M., Mullen R. T., Ohlrogge J. B., Chapman K. D. Plant Physiol. 2013;162:1926–1936. doi: 10.1104/pp.113.222455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T. L., Takano Y., Shimada T., Fujiwara M., Fukao Y., Mori M., Okazaki Y., Saito K., Sasaki R., Aoki K., Hara-Nishimura I. Plant Physiol. 2014;164:105–118. doi: 10.1104/pp.113.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T. L., Hayashi M., Hara-Nishimura I. Plant Physiol. 2018;176:199–207. doi: 10.1104/pp.17.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Esaki T., Kenmoku H., Koeduka T., Kiyoyama Y., Masujima T., Asakawa Y., Matsui K. Phytochemistry. 2016;130:77–84. doi: 10.1016/j.phytochem.2016.06.008. [DOI] [PubMed] [Google Scholar]
- Kumar S., Kempinski C., Zhuang X., Norris A., Mafu S., Zi J., Bell S. A., Nybo S. E., Kinison S. E., Jiang Z., Goklany S., Linscott K. B., Chen X., Jia Q., Brown S. D., Bowman J. L., Babbitt P. C., Peters R. J., Chen F., Chappell J. Plant Cell. 2016;28:2632–2650. doi: 10.1105/tpc.16.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. J. Nat. Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- Pateraki I., Andersen-Ranberg J., Hamberger B., Heskes A. M., Martens H. J., Zerbe P., Bach S. S., Møller B. L., Bohlmann J., Hamberger B. Plant Physiol. 2014;164:1222–1236. doi: 10.1104/pp.113.228429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B. M., Fischedick J. T., Lange M. F., Srividya N., Samec D., Poirier B. C. Plant Physiol. 2017;173:456–469. doi: 10.1104/pp.15.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B. M., Turner G. W. Plant Biotechnol. J. 2013;11:2–22. doi: 10.1111/j.1467-7652.2012.00737.x. [DOI] [PubMed] [Google Scholar]
- Goodger J. Q. D., Heskes A. M., Mitchell M. C., King D. J., Neilson E. H., Woodrow I. E. Plant Methods. 2010;6:20. doi: 10.1186/1746-4811-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heskes A. M., Goodger J. Q. D., Tsegay S., Quach T., Williams S. J., Woodrow I. E. PLoS One. 2012;7:e40856. doi: 10.1371/journal.pone.0040856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L. Mol. Syst. Biol. 2007;3:158. doi: 10.1038/msb4100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen-Ranberg J., Kongstad K. T., Nafisi M., Staerk D., Okkels F. T., Mortensen U. H., Møller B. L., Frandsen R. J. N., Kannangara R. ChemBioChem. 2017;18:1893–1897. doi: 10.1002/cbic.201700331. [DOI] [PubMed] [Google Scholar]
- Hansen E. H., Møller B. L., Kock G. R., Bunner C. M., Kristensen C., Jensen O. R., Okkels F. T., Olsen C. E., Motawia M. S., Hansen J. Appl. Environ. Microbiol. 2009;75:2765–2774. doi: 10.1128/AEM.02681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignea C., Ioannou E., Georgantea P., Loupassaki S., Trikka F. A., Kanellis A. K., Makris A.A. M.M., Roussis V., Kampranis S. C. Metab. Eng. 2015;28:91–103. doi: 10.1016/j.ymben.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Osbourn A. Trends Genet. 2010;26:449–457. doi: 10.1016/j.tig.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Faulkner M., Rodriguez-Ramos J., Dykes G. F., Owen S. V., Casella S., Simpson D. M., Beynon R. J., Liu L. N. Nanoscale. 2017;9:10662–10673. doi: 10.1039/c7nr02524f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates T. O., Kerfeld C. A., Heinhorst S., Cannon G. C., Shively J. M. Nat. Rev. Microbiol. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- Schirrmeister B. E., Sanchez-Baracaldo P., Wacey D. Int. J. Astrobiol. 2016;15:187–204. [Google Scholar]
- You C., Zhang Y. H. ACS Synth. Biol. 2013;2:102–110. doi: 10.1021/sb300068g. [DOI] [PubMed] [Google Scholar]
- Singleton C., Howard T. P., Smirnoff N. J. Exp. Bot. 2014;65:1947–1954. doi: 10.1093/jxb/eru050. [DOI] [PubMed] [Google Scholar]
- Good M. C., Zalatan J. G., Lim W. A. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J., Park S. H. Sci. Signaling. 2015;8:ra66. doi: 10.1126/scisignal.aab3397. [DOI] [PubMed] [Google Scholar]
- Wang Y., Yu O. J. Biotechnol. 2012;157:258–260. doi: 10.1016/j.jbiotec.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Elani Y., Law R. V., Ces O. Nat. Commun. 2014;5:5305. doi: 10.1038/ncomms6305. [DOI] [PubMed] [Google Scholar]
- Fujiwara K., Adachi T., Doi N. ACS Synth. Biol. 2018;7:363–370. doi: 10.1021/acssynbio.7b00365. [DOI] [PubMed] [Google Scholar]
- Jensen M. K., Keasling J. D. FEMS Yeast Res. 2015;15:1–10. doi: 10.1111/1567-1364.12185. [DOI] [PubMed] [Google Scholar]
- Farhi M., Marhevka E., Masci T., Marcos E., Eyal Y., Ovadis M., Abeliovich H., Vainstein A. Metab. Eng. 2011;13:474–481. doi: 10.1016/j.ymben.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Giessen T. W., Silver P. A. J. Mol. Biol. 2016;428:916–927. doi: 10.1016/j.jmb.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Bonacci W., Teng P. K., Afonso B., Niederholtmeyer H., Grob P., Silver P. A., Savage D. F. Proc. Natl. Acad. Sci. U. S. A. 2012;109:478–483. doi: 10.1073/pnas.1108557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. L., Zhu J., Wheeldon I. ACS Synth. Biol. 2017;6:1534–1544. doi: 10.1021/acssynbio.7b00041. [DOI] [PubMed] [Google Scholar]
- Zhao C., Kim Y., Zeng Y., Li M., Wang X., Hu C., Gorman C., Dai S. Y., Ding S. Y., Yuan J. S. ACS Synth. Biol. 2018;7:774–781. doi: 10.1021/acssynbio.7b00368. [DOI] [PubMed] [Google Scholar]
- Roodbeen R., van Hest J. C. Bioessays. 2009;31:1299–1308. doi: 10.1002/bies.200900106. [DOI] [PubMed] [Google Scholar]
- Gallage N. J., Møller B. L. Mol. Plant. 2015;8:40–57. doi: 10.1016/j.molp.2014.11.008. [DOI] [PubMed] [Google Scholar]
- Paddon C. J., Westfall P. J., Pitera D. J., Benjamin K., Fisher K., McPhee D., Leavell M. D., Tai A., Main A., Eng D., Polichuk D. R., Teoh K. H., Reed D. W., Treynor T., Lenihan J., Fleck M., Bajad S., Dang G., Dengrove D., Diola D., Dorin G., Ellens K. W., Fickes S., Galazzo J., Gaucher S. P., Geistlinger T., Henry R., Hepp M., Horning T., Iqbal T., Jiang H., Kizer L., Lieu B., Melis D., Moss N., Regentin R., Secrest S., Tsuruta H., Vazquez R., Westblade L. F., Xu L., Yu M., Zhang Y., Zhao L., Lievense J., Covello P. S., Keasling J. D., Reiling K. K., Renninger N. S., Newman J. D. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]