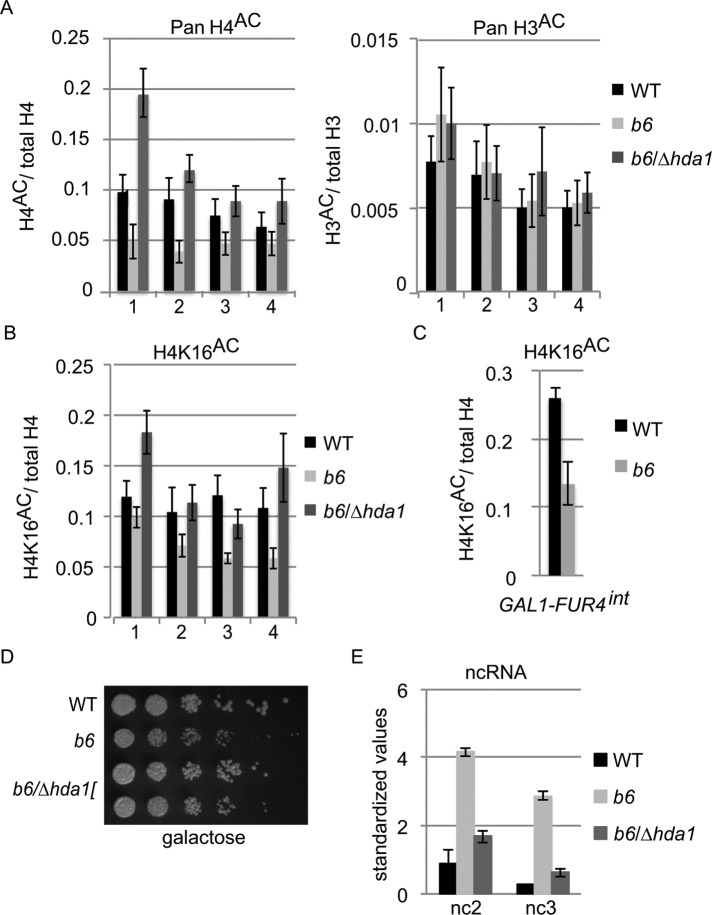
FIGURE 5:
Aberrant transcription at FUR4 and GAL7 correlates with hypoacetylation at histone H4K16. (A, B) ChIP ratios of (A) pan-acetylated histone H4/total H4 pan-acetylated histone H3/total H3, and (B) lysine 16–acetylated H4 in WT (wild type, yDBK165), b6 (brr6-1, yDBK166), and b6/∆hda1 (brr6-1/∆hda1, yDBK169) cells following 2 h galactose induction. DNA was amplified using primer sets #1-4: #1-GAL10 3′ ORF, #2-GAL7 5′ ORF, #3-GAL7 3′ ORF, and #4-GAL7 3′ UTR. Total H4 and H3 values were normalized against WT prior to calculation of ratios. All antibody ChIP levels were >10× above mock ChIP. Error bars (SEM) reflect average of four biological replicates. (C) ChIP ratios of lysine 16–acetylated H4 amplified with primers for the GAL1-FUR4 intergenic region. (D) Growth of WT (wild type, yDBK165), b6 (brr6-1, yDBK166), and b6/∆hda1 (brr6-1/∆hda1, yDBK169) double mutants on YEP +2% galactose, 0.04% sucrose at 30°C. (E) DN9 RT-qPCR detection of GAL ncRNA (nc2 and nc3) transcripts in WT, b6, and b6/∆hda1 following galactose induction for 2 h at 30°C. qPCR data were normalized as in Figure 1. Error bars (SEM) reflect ≥3 biological replicates.