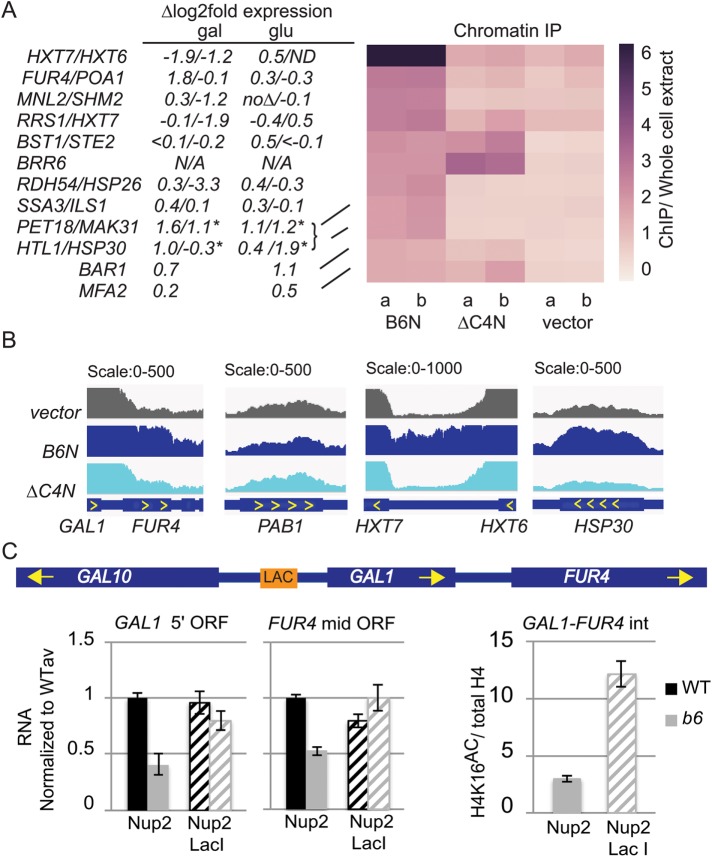
FIGURE 7:
The NLS-Brr6N fragment associates with chromatin at FUR4 and several glucose- and heatshock-responsive genes. (A) Right: Heatmap of the average read density (RPKM, ChIP normalized to whole cell extract) of Brr6 binding sites in ChIP samples from W303 cells carrying vector (pJL602), B6N (pPGAL_NLS-BRR6N-FLAG), or ∆C4N (pPGAL_NLS-brr6∆C4N-FLAG) prepared following O/N galactose induction (two biological replicates). Regions of Brr6 binding were identified by sliding window (see Materials and Methods). Only regions with at least 1.5-fold ChIP/WCE are shown. Values are shown in Supplemental Table S5. qPCR confirming absence of CHIII disomy following B6N expression is shown in Supplemental Figure S7. (A) Left, changes in expression in brr6-1 obtained by RNAseq analysis are shown for associated genes (*genes on CHIII not affected by brr6-1 are predicted to show ∆log2 ≈ 1 expression changes due to disomy). (B) Bar plot of RPKM-normalized reads enriched in vector (pJL602), B6N (pPGAL_NLS-BRR6N-FLAG), or ∆C4N (pPGAL_NLS-brr6∆C4N-FLAG) samples showing GAL1-FUR4, PAB1, and HXT7-HXT6 and HSP30. (C) Left: RT-qPCR analyses of GAL1 5′ ORF and FUR4 mid-ORF transcript levels in cells carrying a LAC O tag upstream of GAL1 (see schematic) and either Nup2 and Nup2-Lac I URA3-marked constructs. (C) Right, comparison of H4K16 acetylation in the FUR4-GAL1 intergenic region brr6-1 cells carrying the LAC O tag and the Nup2 vs. Nup2-Lac I construct. Error bars (SEM) reflect three biological replicates.